![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-00-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
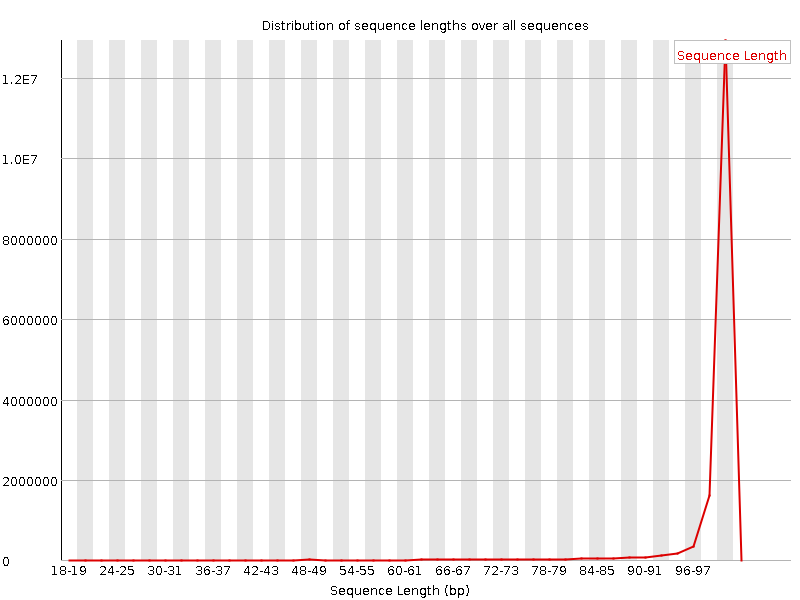
| Total Sequences | 16351272 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
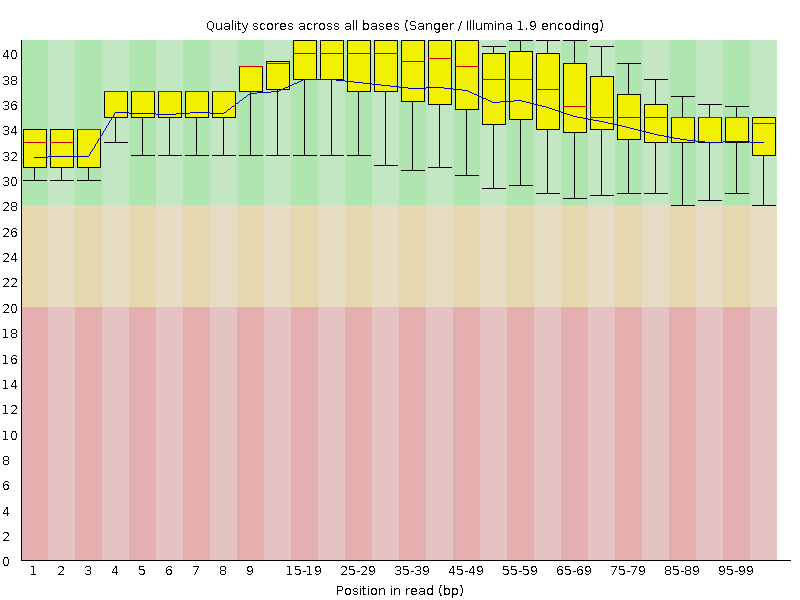
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

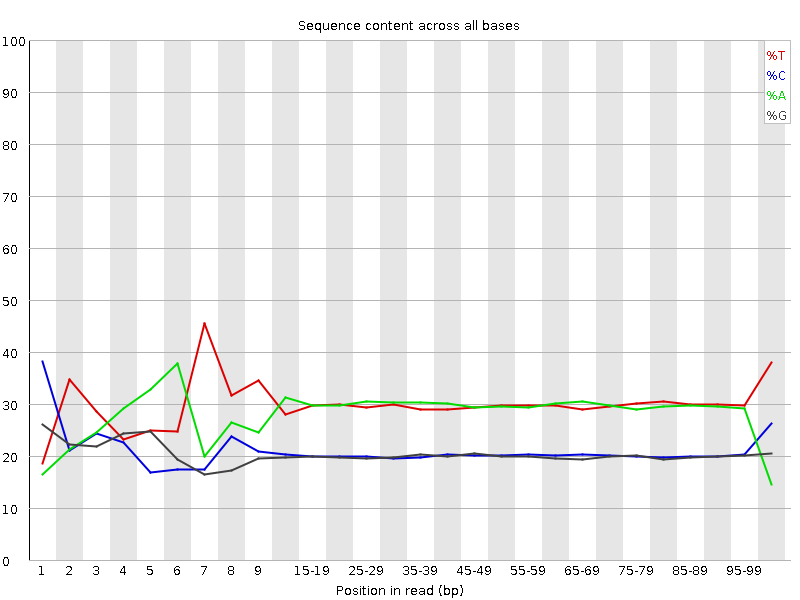
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
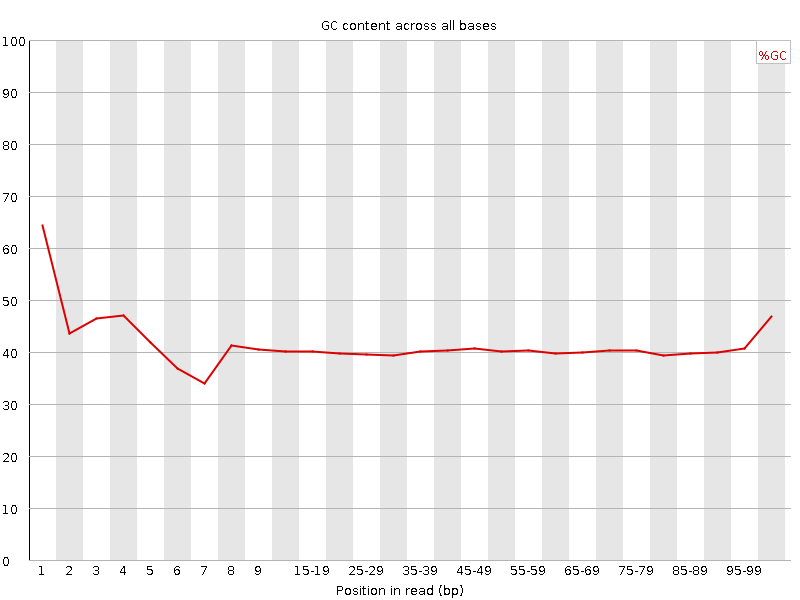
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
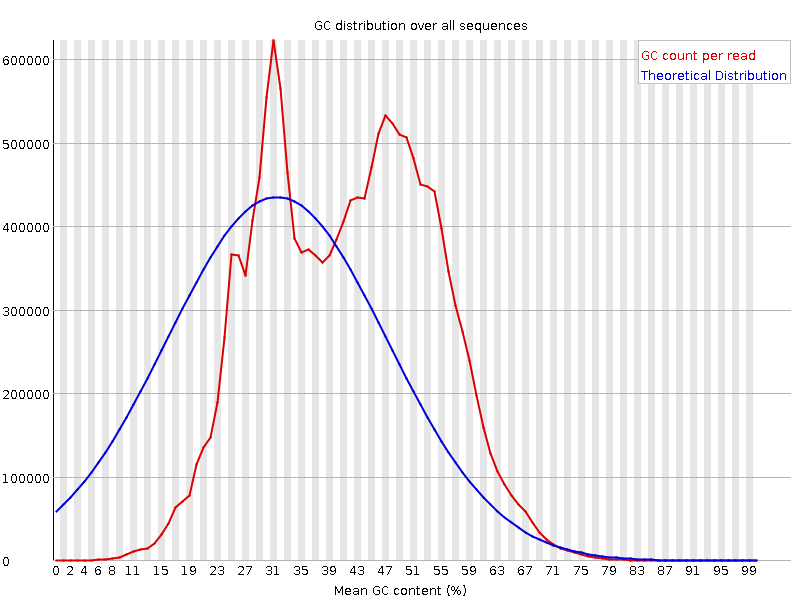
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels

![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 71419 | 0.43677947501576636 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 67212 | 0.4110505898256723 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 50900 | 0.3112907668589942 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 46042 | 0.2815805400338273 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 30846 | 0.1886458741558455 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 27308 | 0.16700841377967415 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 26812 | 0.1639750106291425 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 23106 | 0.1413101072503717 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 18459 | 0.11289029991061246 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 17118 | 0.10468910308629199 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 17064 | 0.10435885354974218 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 16896 | 0.10333141054714276 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
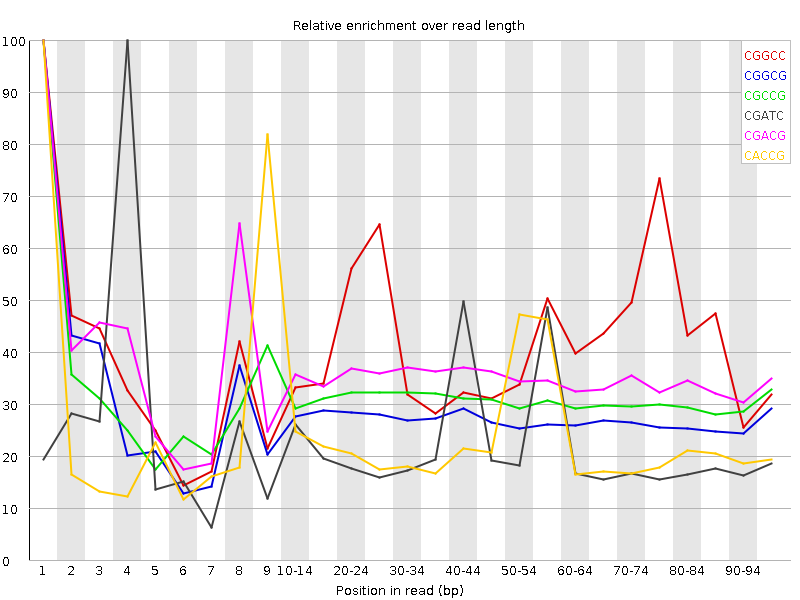
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1368445 | 2.544987 | 6.0949903 | 1 |
| CGGCG | 1268060 | 2.4004433 | 8.682649 | 1 |
| CGCCG | 1275205 | 2.3715825 | 7.627036 | 1 |
| CGATC | 2417930 | 2.1102903 | 9.486108 | 4 |
| CGACG | 1574665 | 2.037099 | 5.7362194 | 1 |
| CACCG | 1597595 | 2.0304728 | 8.606954 | 1 |
| CCTCG | 1499760 | 1.8817183 | 5.5144186 | 1 |
| CTGGC | 1420725 | 1.8144137 | 7.2438455 | 1 |
| CGCGG | 950565 | 1.799424 | 5.932759 | 1 |
| CCGCG | 955025 | 1.7761227 | 5.9229193 | 1 |
| CCGGC | 953345 | 1.7729985 | 6.697677 | 1 |
| GCCGA | 1367095 | 1.7685715 | 5.3339972 | 1 |
| CCGAT | 1981025 | 1.7289739 | 7.9335837 | 3 |
| ACCGA | 1760705 | 1.5566206 | 5.7897396 | 2 |
| CAGCG | 1149010 | 1.4864414 | 5.1468616 | 1 |
| CTTTC | 2516230 | 1.4626005 | 6.6756163 | 1 |
| TCCTT | 2400495 | 1.3953276 | 5.7506003 | 2 |
| GATCT | 2310565 | 1.3847924 | 6.2790723 | 5 |
| GACCT | 1582280 | 1.3809625 | 5.1952996 | 95-97 |
| GGGGC | 661635 | 1.2748634 | 6.4997387 | 95-97 |
| CCTTT | 2180090 | 1.2672136 | 5.8403625 | 3 |
| CTCGG | 985080 | 1.2580496 | 5.648964 | 1 |
| TTTCG | 1980320 | 1.1716669 | 5.928693 | 5 |
| TTCGT | 1936985 | 1.1460276 | 5.950171 | 6 |
| CGTAC | 1206730 | 1.0531945 | 7.3622174 | 5 |
| GTCCT | 1103695 | 0.9509329 | 7.5439787 | 1 |
| GTACA | 1559105 | 0.94654113 | 5.832239 | 6 |
| TCGTA | 1572100 | 0.9422077 | 5.8682327 | 7 |