![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-03-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17038723 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 38 |
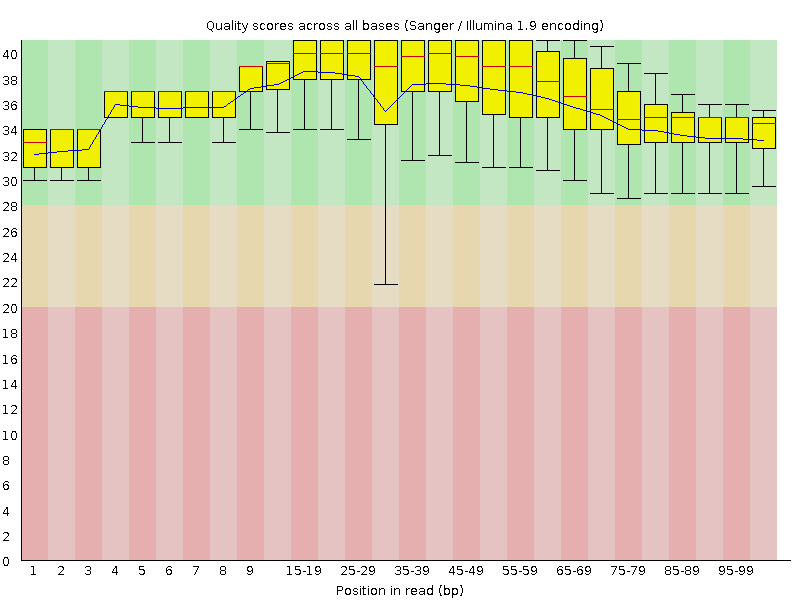
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
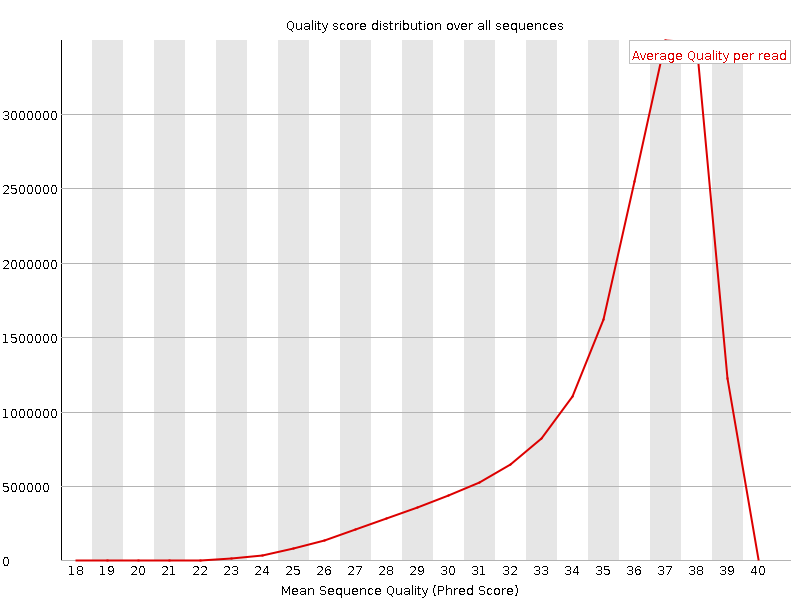
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

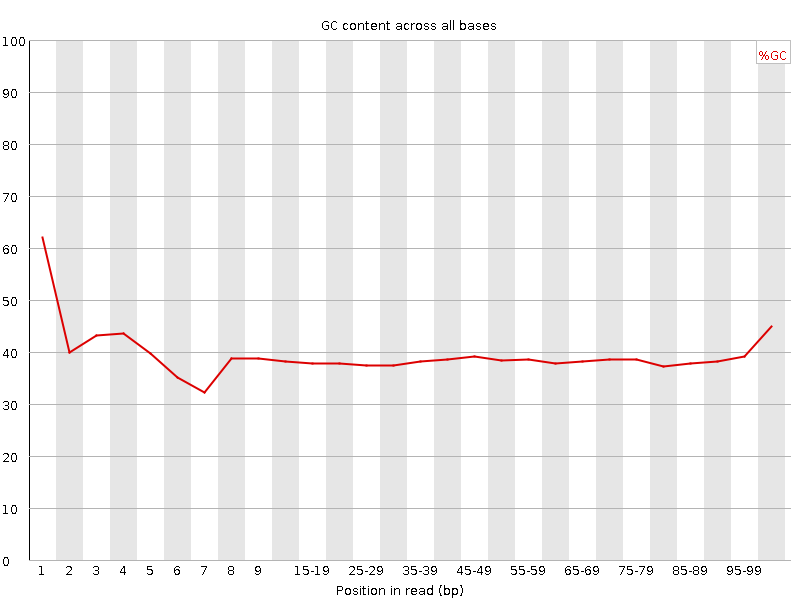
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
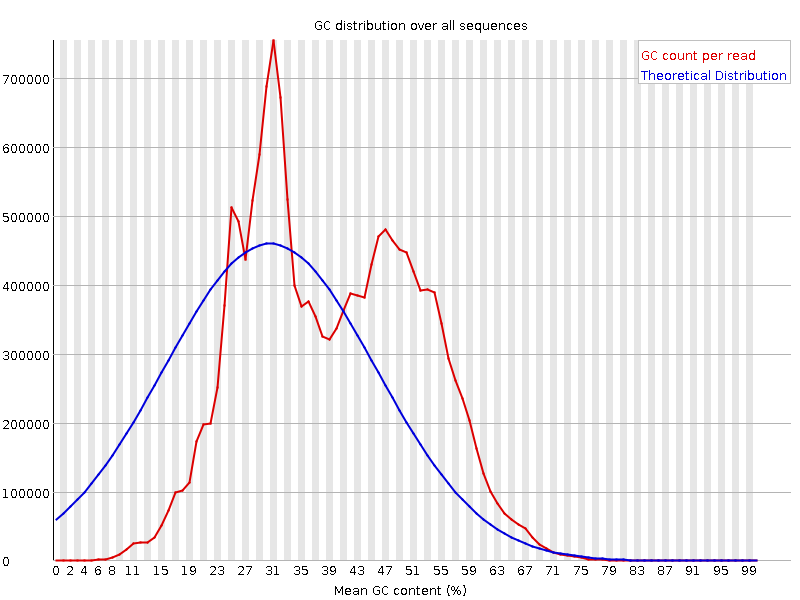
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
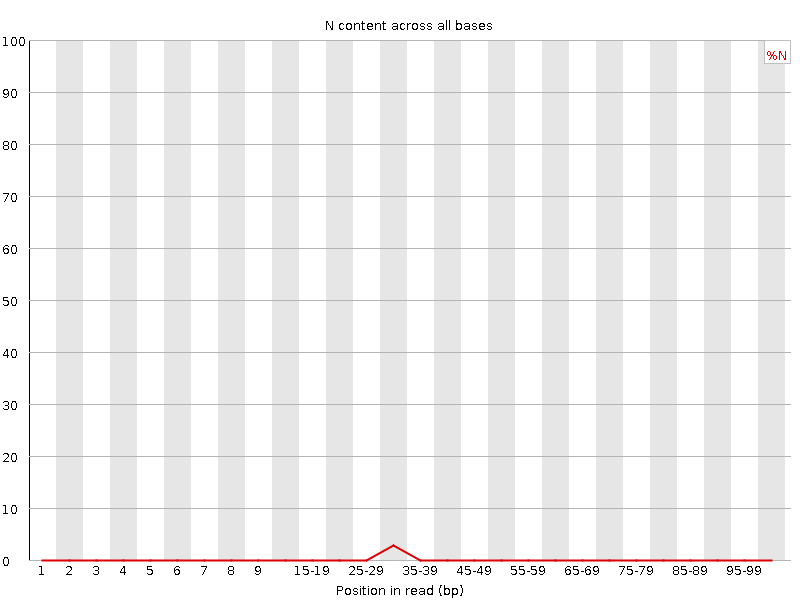
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
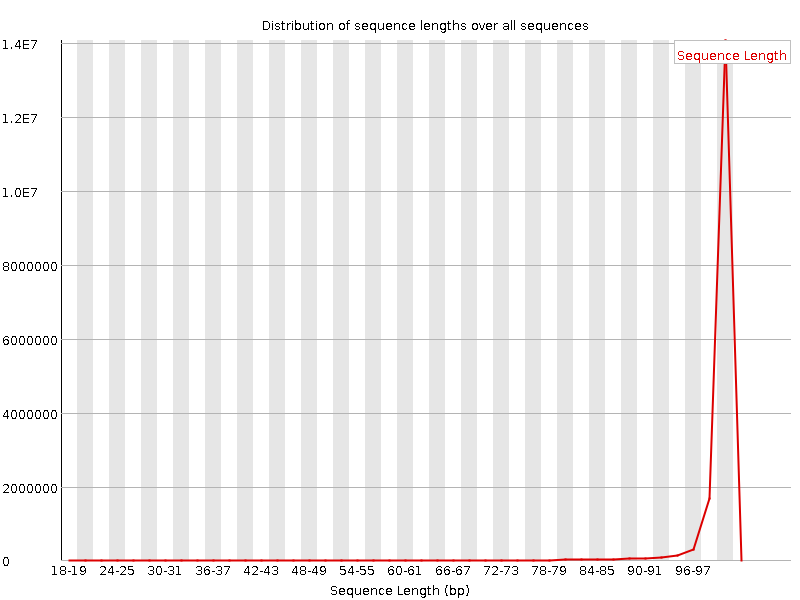
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 90665 | 0.5321114733774357 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 70905 | 0.41614034103377345 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 68312 | 0.40092206440588296 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 52212 | 0.3064314150772919 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 41941 | 0.24615107599319502 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 38645 | 0.2268069033107704 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 36162 | 0.21223421497021813 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 30700 | 0.18017782201166133 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 26844 | 0.15754701804824223 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 25580 | 0.15012862172828326 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 24820 | 0.14566819356121935 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 23860 | 0.14003396850808597 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 22466 | 0.13185260421218187 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 19889 | 0.11672823133517694 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 19726 | 0.11577158687303034 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 19229 | 0.11285470161114773 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 19012 | 0.11158113198976238 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 18531 | 0.10875815047876533 | No Hit |
| GGCAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATA | 18151 | 0.10652793639523336 | No Hit |
| GTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATG | 17441 | 0.1023609574496868 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
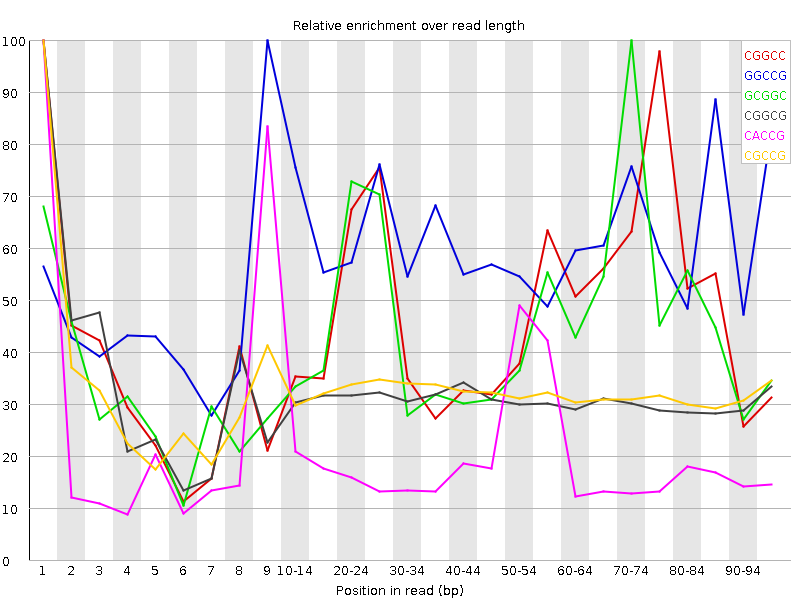
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1543695 | 3.502497 | 7.3243523 | 1 |
| GGCCG | 1465220 | 3.3478646 | 5.529268 | 9 |
| GCGGC | 1421515 | 3.2480035 | 7.2044497 | 70-74 |
| CGGCG | 1081610 | 2.4713583 | 7.9015694 | 1 |
| CACCG | 1660540 | 2.381777 | 11.829029 | 1 |
| CGCCG | 1041970 | 2.3641305 | 7.31149 | 1 |
| GGTCG | 1607375 | 2.3269625 | 5.6243896 | 85-89 |
| CTGGC | 1596240 | 2.2946775 | 10.059439 | 1 |
| CGATC | 2506500 | 2.2778578 | 10.913214 | 4 |
| CCTCG | 1590360 | 2.270231 | 6.817838 | 1 |
| GCCGG | 956540 | 2.1855872 | 5.0705605 | 1 |
| CCGGC | 961155 | 2.1807692 | 7.688748 | 1 |
| GGCTT | 2216090 | 2.01845 | 6.624355 | 3 |
| CCGAT | 2134420 | 1.9397188 | 9.571077 | 3 |
| GACCT | 2109885 | 1.9174219 | 7.8232737 | 95-97 |
| CGACG | 1313340 | 1.8970453 | 5.478226 | 1 |
| GGGGC | 811780 | 1.8678936 | 11.428573 | 95-97 |
| AAGCC | 2029000 | 1.8527554 | 5.0476685 | 5 |
| TGGCT | 2030610 | 1.8495117 | 5.7379003 | 45-49 |
| GCAAA | 3141740 | 1.8263779 | 5.4927616 | 2 |
| CGCGG | 793765 | 1.8136644 | 5.3813996 | 1 |
| GCCGA | 1252405 | 1.8090283 | 5.349953 | 1 |
| CCGCG | 794215 | 1.801998 | 5.1465516 | 1 |
| ACCGA | 1900975 | 1.735851 | 7.6565413 | 2 |
| CAGCG | 1183895 | 1.7100693 | 7.109617 | 1 |
| ATATT | 6935735 | 1.5995629 | 5.2565947 | 4 |
| CGCAA | 1711890 | 1.5631906 | 5.278239 | 1 |
| GTCGC | 1062090 | 1.5268089 | 6.2194586 | 95-97 |
| CTTTC | 2634130 | 1.4989164 | 9.121764 | 1 |
| GATCT | 2504200 | 1.4418999 | 6.928332 | 5 |
| CGAAC | 1562070 | 1.4263843 | 5.459721 | 90-94 |
| TCCTT | 2486860 | 1.4151144 | 6.707512 | 2 |
| CCTTT | 2473950 | 1.4077681 | 6.7152934 | 3 |
| CTAAA | 3768315 | 1.3782437 | 5.702301 | 8 |
| CAAAT | 3491030 | 1.2768278 | 5.3889365 | 9 |
| AGACC | 1363280 | 1.2448617 | 6.2719116 | 95-97 |
| GCGTA | 1341710 | 1.2279094 | 5.017279 | 1 |
| CAGAC | 1318850 | 1.2042911 | 5.9437017 | 95-97 |
| CTCGG | 834295 | 1.199342 | 5.675495 | 1 |
| CTCGT | 1325665 | 1.1989902 | 5.0199304 | 1 |
| ACACC | 1320005 | 1.1969137 | 5.356975 | 8 |
| CTCAT | 2032095 | 1.1618801 | 6.5619073 | 1 |
| TTTCG | 1904495 | 1.091362 | 7.478933 | 2 |
| CGTAC | 1188940 | 1.0804852 | 10.980921 | 5 |
| TTCGT | 1873140 | 1.0733942 | 7.368147 | 3 |
| CTCCC | 728785 | 1.0330594 | 5.5608797 | 1 |
| GTCCT | 1008545 | 0.9121728 | 9.428497 | 1 |
| TCGTA | 1525665 | 0.8784667 | 6.9769325 | 4 |
| GTACA | 1498315 | 0.8668548 | 7.541268 | 6 |
| TACAA | 2106675 | 0.77050656 | 5.414461 | 7 |