![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-07-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17635978 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
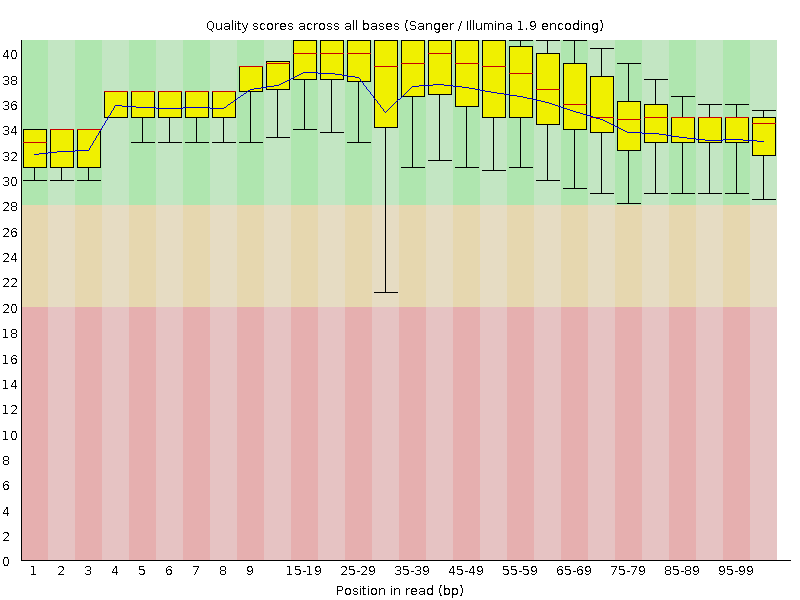
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
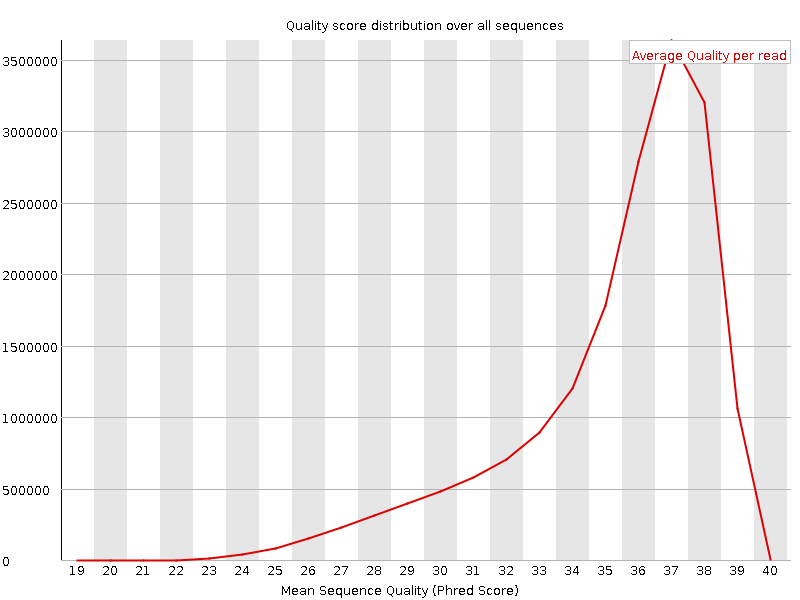
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

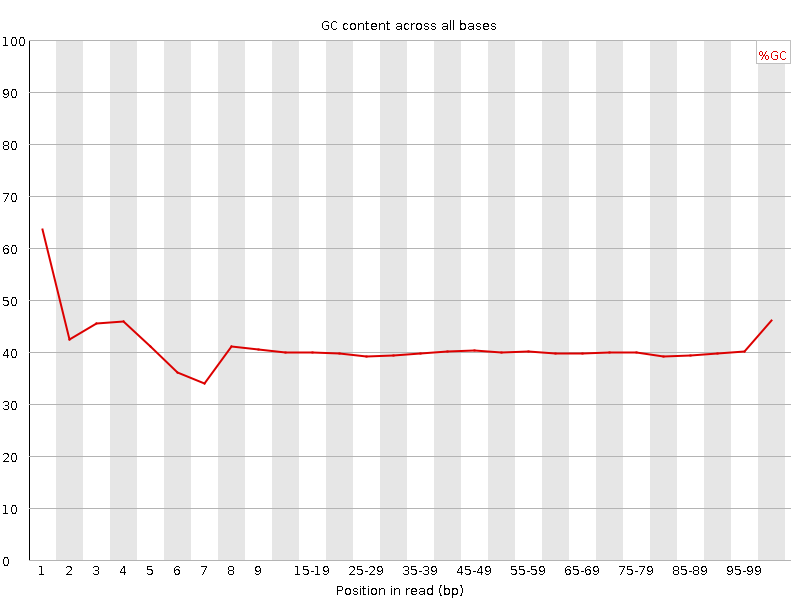
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
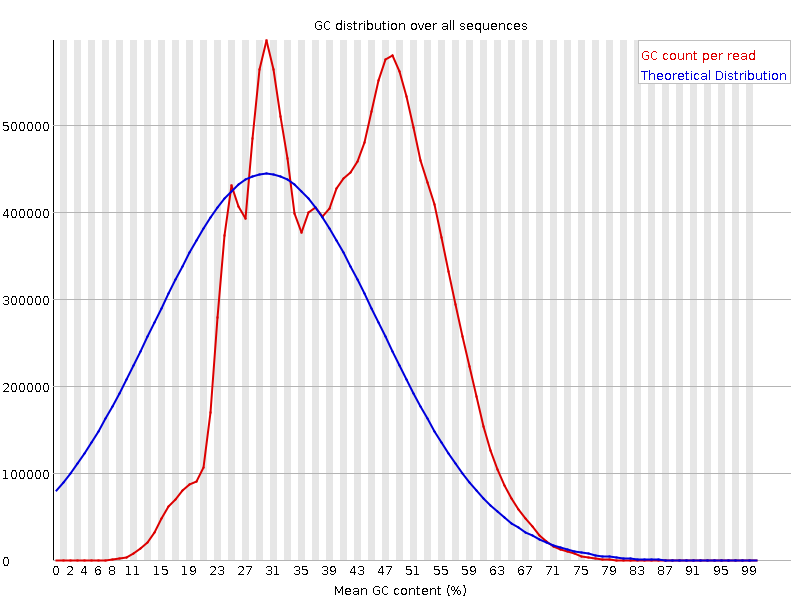
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
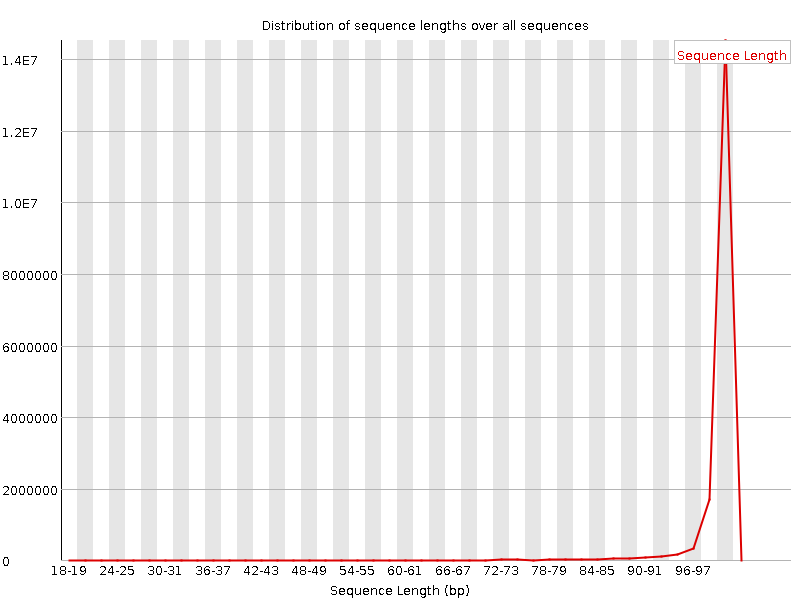
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
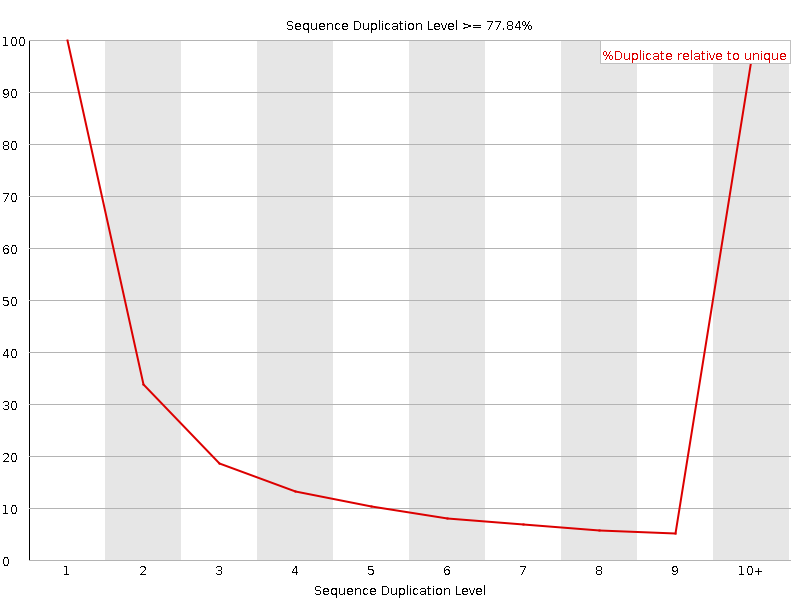
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAACACTAATTATAGATAGAAACCGATCTGGCT | 61181 | 0.3469101628500557 | No Hit |
| GTCCTTTCGTACAAATAATTTAACACTAATTATAGATAGAAACCGATCTG | 52036 | 0.29505593622310033 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 49862 | 0.28272886255585034 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 28395 | 0.16100609787560405 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 26861 | 0.1523079695381793 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 24289 | 0.13772414549394427 | No Hit |
| CAAATATTGAGCTCAACTGTATATTAAAAACATAGCTTTTAGATTATAAT | 23952 | 0.13581327896870818 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 19943 | 0.1130813386136 | No Hit |
| TAAAAACATAGCTTTTAGATTATAATTTAAGGTTATTTCTGCCCTATGAA | 18139 | 0.1028522489651552 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 17844 | 0.10117953197718892 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 17716 | 0.10045374291122386 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
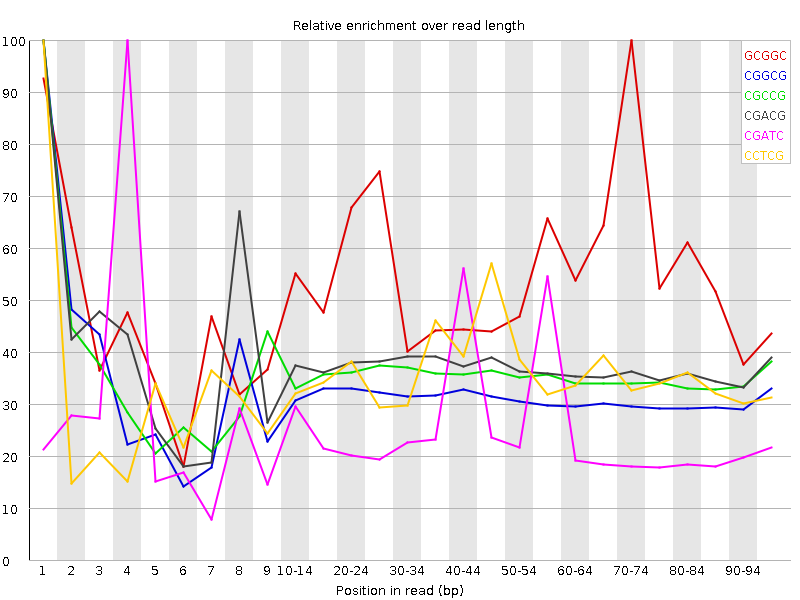
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCGGC | 1531410 | 2.764766 | 5.0528564 | 70-74 |
| CGGCG | 1435975 | 2.5924702 | 8.209244 | 1 |
| CGCCG | 1414955 | 2.526823 | 7.1106815 | 1 |
| CGACG | 1699555 | 2.0736585 | 5.542266 | 1 |
| CGATC | 2416660 | 1.9784824 | 7.797574 | 4 |
| CCTCG | 1587275 | 1.9019496 | 5.2908196 | 1 |
| CGCGG | 1050030 | 1.8956953 | 5.6321406 | 1 |
| CACCG | 1560965 | 1.8839117 | 6.6024137 | 1 |
| CCGCG | 1044670 | 1.8655691 | 5.700006 | 1 |
| CCGGC | 957845 | 1.7105172 | 6.1280966 | 1 |
| ATATT | 6579035 | 1.6749917 | 5.129369 | 4 |
| CTGGC | 1343345 | 1.6273054 | 6.6075616 | 1 |
| CCGAT | 1986315 | 1.6261657 | 6.1480436 | 3 |
| CAGCG | 1259990 | 1.5373373 | 5.076477 | 1 |
| CTTTC | 2702770 | 1.4740759 | 6.9488673 | 1 |
| GACCT | 1753650 | 1.4356865 | 5.2753835 | 95-97 |
| GTCGG | 1097625 | 1.3442193 | 5.135254 | 1 |
| TCCTT | 2435090 | 1.3280845 | 5.166612 | 2 |
| GATCT | 2280510 | 1.2664788 | 5.0649595 | 5 |
| CCTTT | 2289960 | 1.2489316 | 5.13167 | 3 |
| CTCGG | 1014710 | 1.2292025 | 5.261926 | 1 |
| TTTCG | 2116970 | 1.1672398 | 5.4190955 | 2 |
| TTCGT | 2005075 | 1.105544 | 5.3066254 | 6 |
| CGTAC | 1256640 | 1.028792 | 7.1596684 | 5 |
| GTCCT | 1170740 | 0.9516046 | 6.6716776 | 1 |
| GTACA | 1658940 | 0.9279343 | 5.6366653 | 6 |
| TCGTA | 1608170 | 0.8930955 | 5.2347507 | 7 |
| CTCCC | 752620 | 0.8920474 | 5.5321236 | 1 |