![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-10-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 18232041 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 39 |
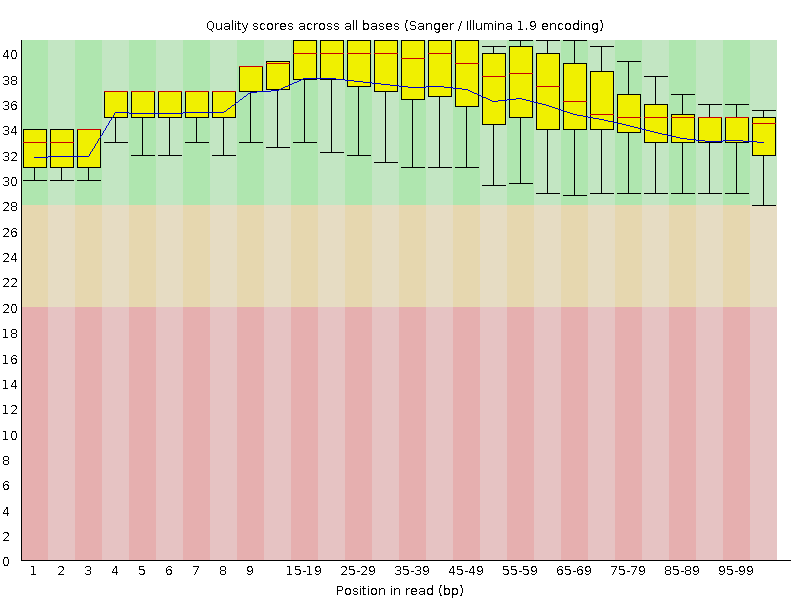
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
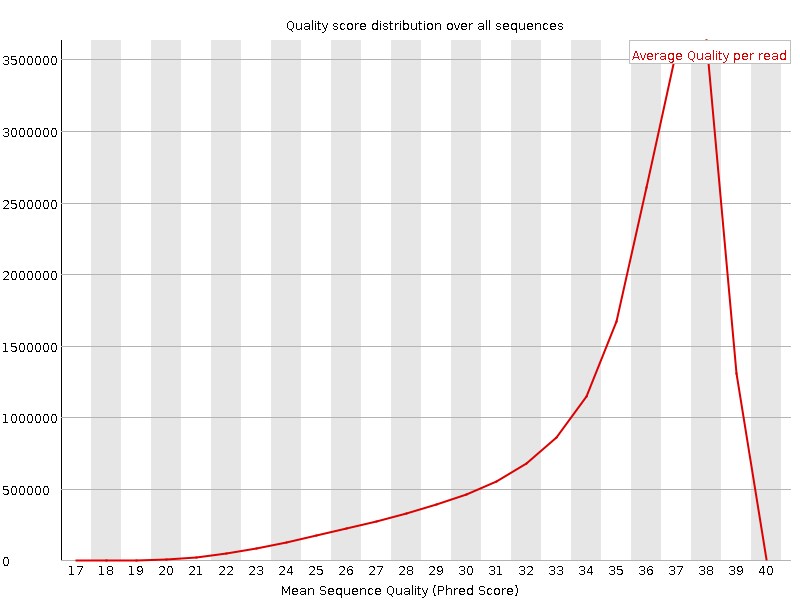
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
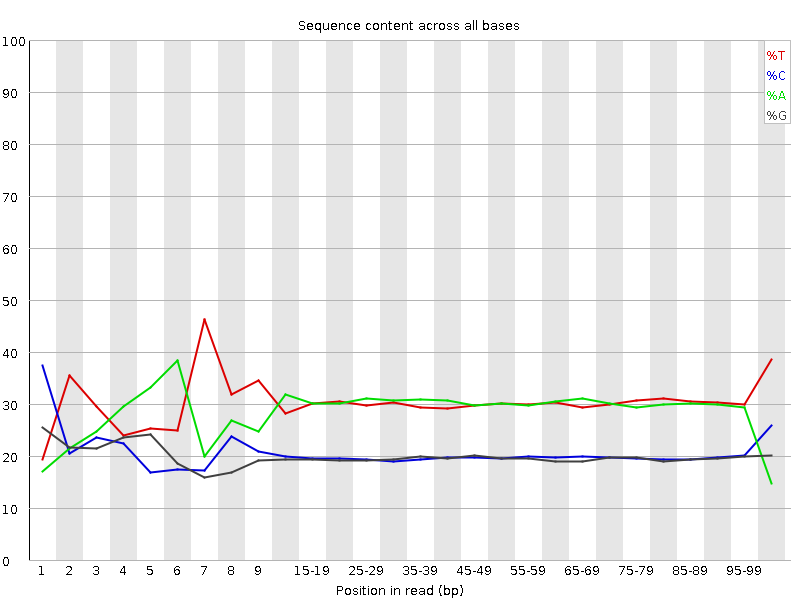
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
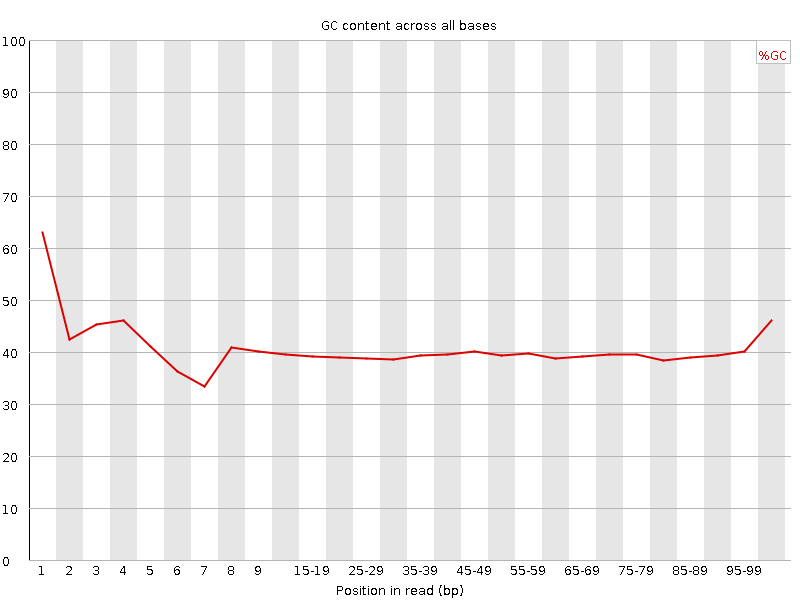
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
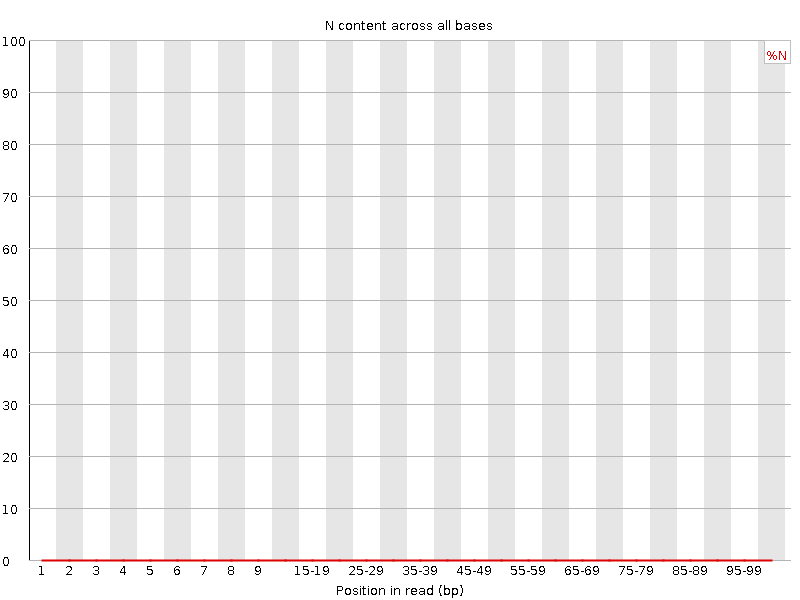
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
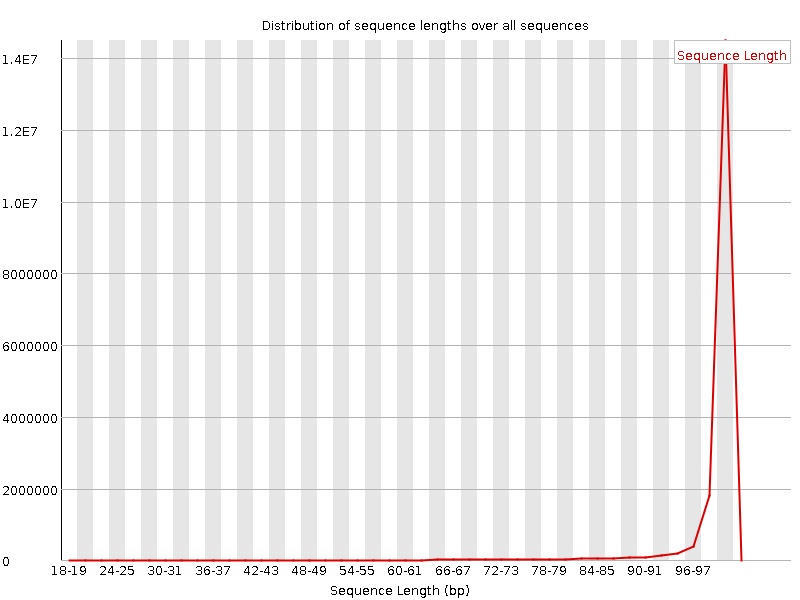
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 92348 | 0.5065148767491253 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 84359 | 0.46269641451552246 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 61970 | 0.3398961202423799 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 61823 | 0.33908984737364295 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 40384 | 0.2215001600753311 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 33940 | 0.18615579023763712 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 33242 | 0.18232736532349836 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 26283 | 0.14415829802050137 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 26055 | 0.14290775234654202 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 24301 | 0.1332873264161703 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 23433 | 0.12852647709600917 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 23318 | 0.12789571940958228 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 21746 | 0.11927353607859921 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 20327 | 0.11149053471303624 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 19499 | 0.10694907937076271 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 19274 | 0.1057149882451449 | No Hit |
| GTCTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATT | 19051 | 0.10449186681842147 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 18798 | 0.10310419990828236 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 18649 | 0.102286957340651 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
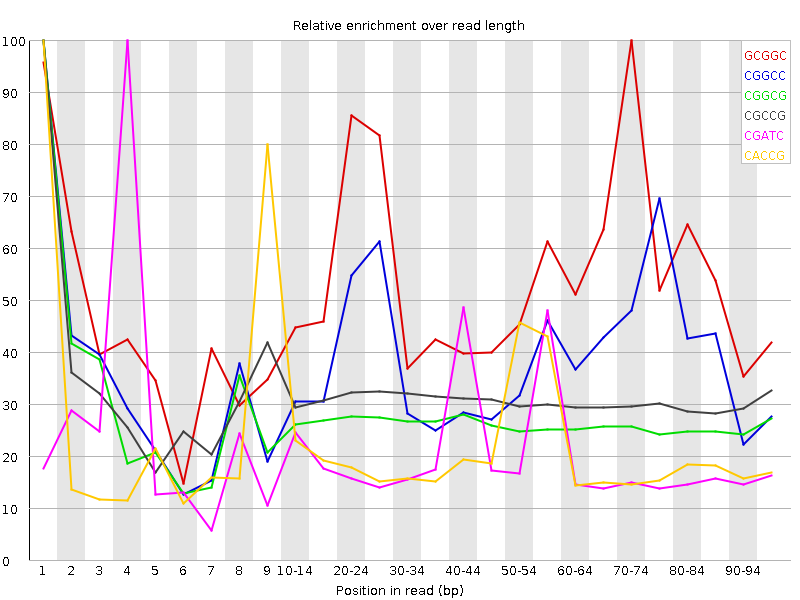
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCGGC | 1546930 | 2.8929508 | 5.3322625 | 70-74 |
| CGGCC | 1542510 | 2.8285966 | 7.283668 | 1 |
| CGGCG | 1337780 | 2.5018141 | 9.324031 | 1 |
| CGCCG | 1362255 | 2.498052 | 8.041611 | 1 |
| CGATC | 2751315 | 2.2188692 | 10.784728 | 4 |
| CACCG | 1820260 | 2.2052786 | 10.310156 | 1 |
| GGTCG | 1709590 | 2.128255 | 5.0023565 | 95-97 |
| CGACG | 1655575 | 2.0455317 | 6.0439463 | 1 |
| CCTCG | 1660890 | 1.9880071 | 6.036413 | 1 |
| CTGGC | 1613380 | 1.9694321 | 8.349347 | 1 |
| CCGGC | 1054670 | 1.9340143 | 7.003429 | 1 |
| CGCGG | 999475 | 1.8691419 | 5.8346643 | 1 |
| CCGCG | 1016605 | 1.864212 | 5.9919753 | 1 |
| CCGAT | 2220225 | 1.790558 | 8.984384 | 3 |
| GCCGA | 1419820 | 1.7542467 | 5.0835843 | 1 |
| ACCGA | 2033880 | 1.6602356 | 6.927492 | 2 |
| GGCTT | 2027570 | 1.647561 | 5.4039807 | 3 |
| GTCGC | 1321465 | 1.6130953 | 5.9177666 | 95-97 |
| CAGCG | 1269100 | 1.5680258 | 5.2654314 | 1 |
| GACCT | 1931345 | 1.5575831 | 6.2132654 | 95-97 |
| GGGGC | 795730 | 1.5176213 | 7.754477 | 95-97 |
| CTTTC | 2860840 | 1.4878731 | 7.497574 | 1 |
| GATCT | 2667105 | 1.4318341 | 6.958833 | 5 |
| TCCTT | 2680550 | 1.3941073 | 6.227093 | 2 |
| CCTTT | 2526865 | 1.3141785 | 6.336494 | 3 |
| CTCGG | 1037290 | 1.2662065 | 5.787603 | 1 |
| CTAAA | 3386810 | 1.21586 | 5.083024 | 8 |
| TTTCG | 2189940 | 1.1615338 | 6.502352 | 5 |
| AGACC | 1398920 | 1.1419243 | 5.246589 | 95-97 |
| TTCGT | 2097520 | 1.1125146 | 6.4719915 | 6 |
| CGTAC | 1333775 | 1.0756574 | 8.563856 | 5 |
| GTCCT | 1204150 | 0.95944244 | 8.515008 | 1 |
| CCGGG | 501415 | 0.9377082 | 5.0467434 | 1 |
| GTACA | 1705475 | 0.92672527 | 6.4330974 | 6 |
| TCGTA | 1704920 | 0.91528565 | 6.4002094 | 7 |