![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-28-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 13905764 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 39 |
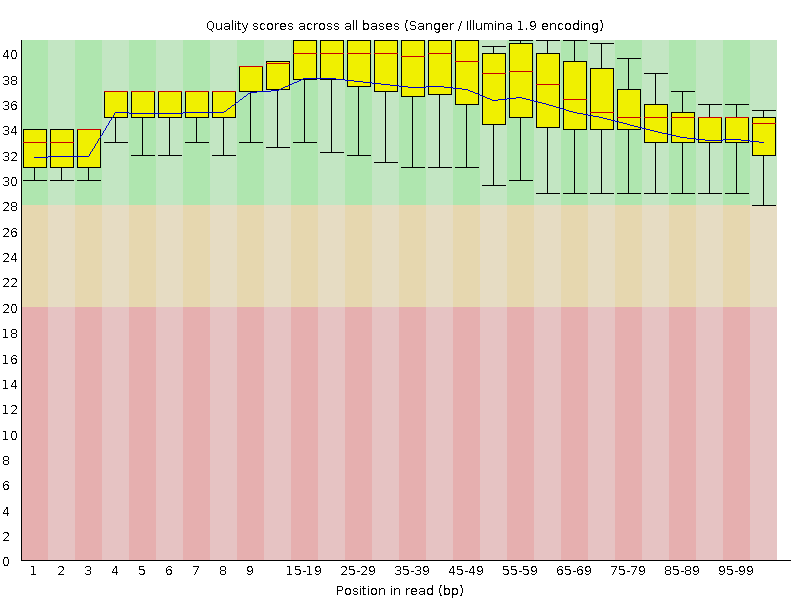
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
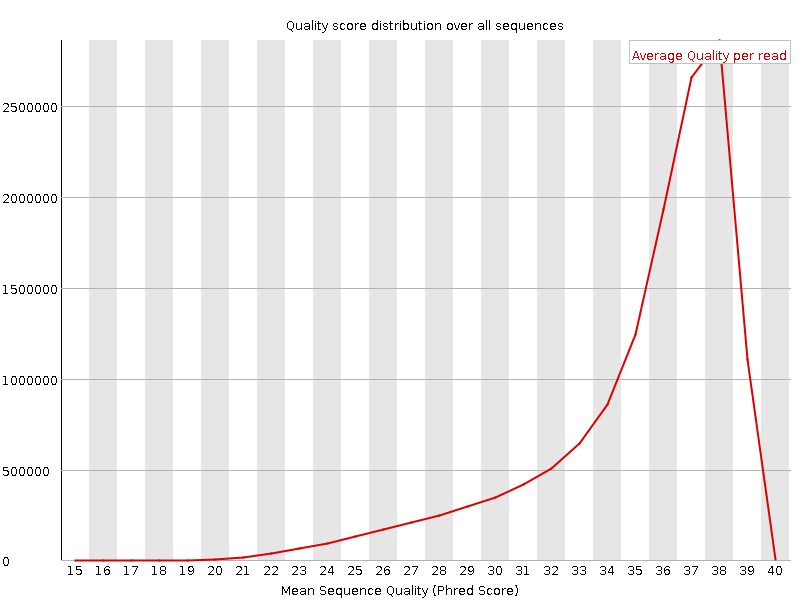
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
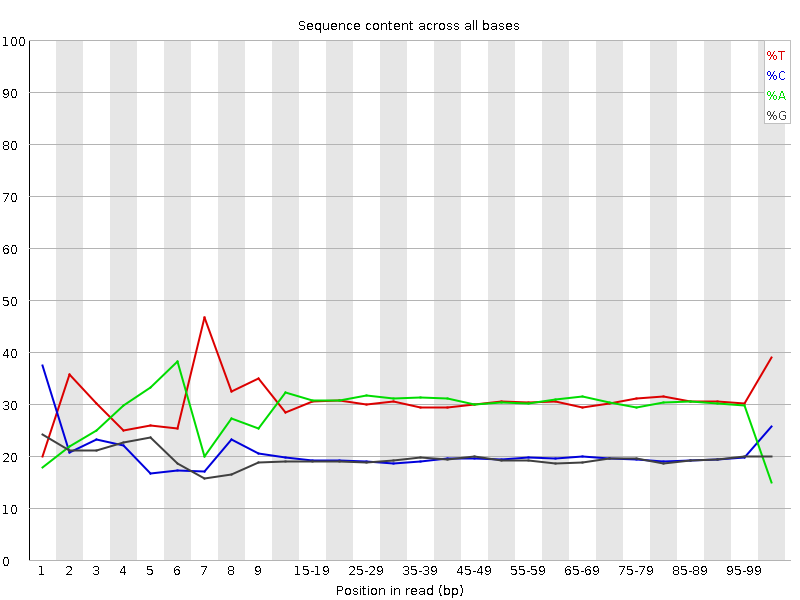
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
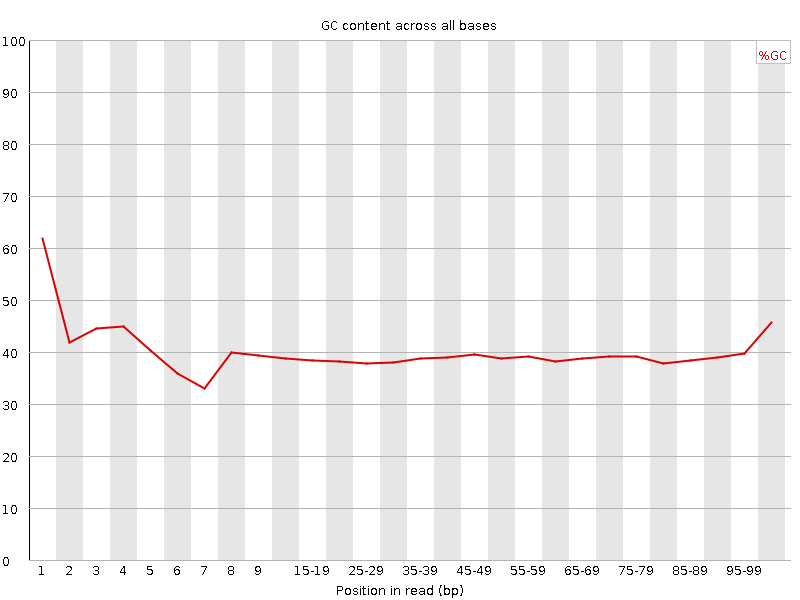
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
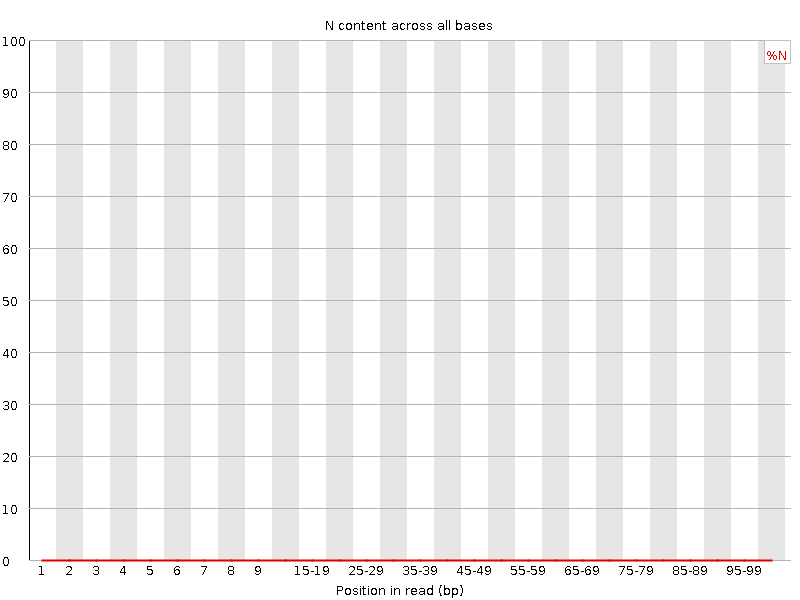
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
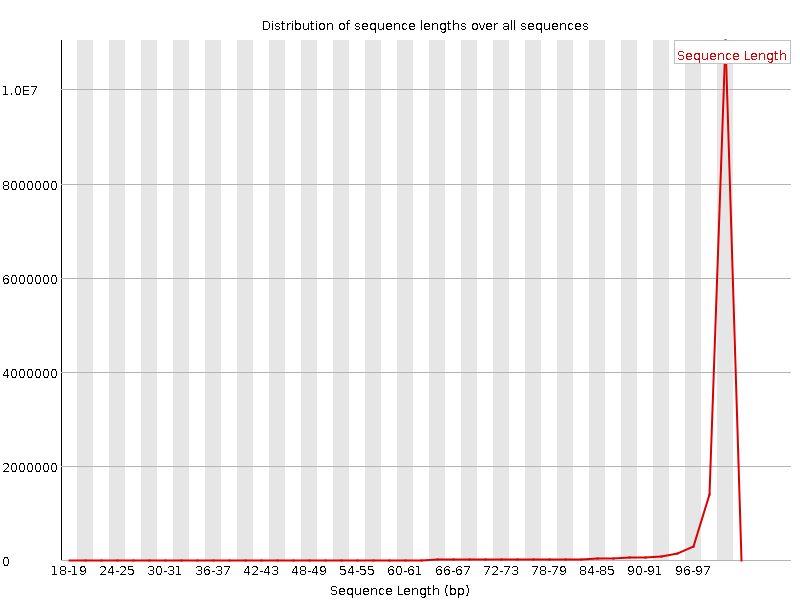
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 89923 | 0.6466599030445217 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 81681 | 0.5873895170376832 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 58359 | 0.41967489164924704 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 57896 | 0.4163453370846794 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 35147 | 0.25275130514224176 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 31062 | 0.22337499759092702 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 30312 | 0.21798155067208103 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 23738 | 0.170706190612756 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 23540 | 0.16928232062618062 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 20597 | 0.14811843491662882 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 20550 | 0.14778044557638115 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 19695 | 0.14163191608889666 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 18789 | 0.13511663221093068 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 18165 | 0.1306292843744508 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 18092 | 0.1301043222076831 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 17474 | 0.12566012194655396 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 16809 | 0.12087793234517716 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 16233 | 0.1167357651115034 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 16143 | 0.11608855148124188 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 15987 | 0.1149667145221219 | No Hit |
| CTCCCAACTAAATATAATTCATATTATTAATGATACAAAAATTTTTAATA | 15247 | 0.1096451802288605 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 14684 | 0.10559649940844675 | No Hit |
| GGCAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATA | 14511 | 0.10435241098583294 | No Hit |
| GTCTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATT | 14352 | 0.10320900023903758 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 14021 | 0.10082869233218686 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
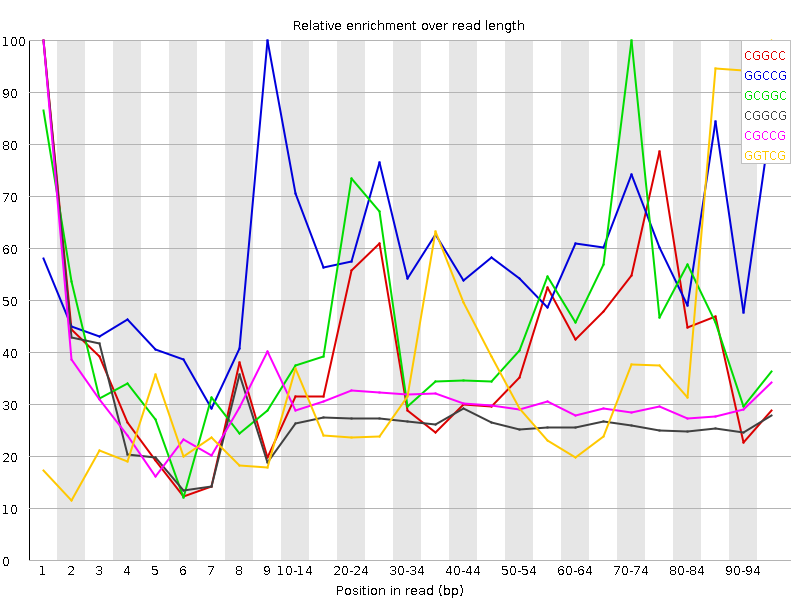
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1264290 | 3.268867 | 7.906194 | 1 |
| GGCCG | 1203345 | 3.1785674 | 5.2843394 | 9 |
| GCGGC | 1177185 | 3.1094673 | 6.574572 | 70-74 |
| CGGCG | 948985 | 2.5066898 | 9.240965 | 1 |
| CGCCG | 954055 | 2.4667432 | 8.052087 | 1 |
| GGTCG | 1424170 | 2.4496918 | 6.201295 | 95-97 |
| CACCG | 1450725 | 2.4128077 | 12.541134 | 1 |
| CGATC | 2183385 | 2.3647017 | 12.022001 | 4 |
| CTGGC | 1340805 | 2.2574835 | 9.280338 | 1 |
| GCCGG | 836750 | 2.2102275 | 5.473781 | 1 |
| CCGGC | 849760 | 2.197085 | 7.6484494 | 1 |
| CCTCG | 1272420 | 2.0970018 | 6.4614997 | 1 |
| CCGAT | 1864490 | 2.0193245 | 10.427276 | 3 |
| GGCTT | 1825005 | 2.000928 | 6.162163 | 3 |
| CGACG | 1149245 | 1.9527242 | 5.6487308 | 1 |
| CGCGG | 704655 | 1.8613061 | 5.8464007 | 1 |
| CCGCG | 718725 | 1.8582894 | 5.868553 | 1 |
| GCCGA | 1075590 | 1.8275744 | 5.6711025 | 1 |
| TGGCT | 1658020 | 1.8178462 | 6.0870523 | 45-49 |
| GACCT | 1674845 | 1.8139305 | 7.900353 | 95-97 |
| ACCGA | 1654110 | 1.8079222 | 8.314451 | 2 |
| GGGGC | 658115 | 1.7759622 | 10.160957 | 95-97 |
| AGGTC | 1595240 | 1.7650733 | 5.156154 | 95-97 |
| CAGCG | 962580 | 1.6355549 | 5.9723144 | 1 |
| GTCGC | 947190 | 1.5947626 | 7.095506 | 95-97 |
| GATCT | 2166315 | 1.5278336 | 7.649653 | 5 |
| CTTTC | 2178345 | 1.4901193 | 8.903881 | 1 |
| TCCTT | 2149045 | 1.4700764 | 7.5882125 | 2 |
| CGAAC | 1341155 | 1.4658662 | 5.871758 | 90-94 |
| CTAAA | 3005725 | 1.376132 | 6.1554484 | 8 |
| CCTTT | 1995385 | 1.3649639 | 7.5146317 | 3 |
| CAAAT | 2810445 | 1.2867256 | 5.512573 | 9 |
| AGACC | 1159775 | 1.2676201 | 6.683014 | 95-97 |
| CTCGG | 725240 | 1.2210704 | 5.6313815 | 1 |
| CAGAC | 1111520 | 1.214878 | 6.1554756 | 95-97 |
| ACACC | 1124780 | 1.2033508 | 5.081194 | 8 |
| TTTCG | 1634550 | 1.1423082 | 7.7651753 | 5 |
| TTCGT | 1627625 | 1.1374687 | 7.82279 | 6 |
| CGTAC | 1046480 | 1.1333838 | 10.885993 | 5 |
| CTCAT | 1594660 | 1.1008593 | 5.7235894 | 1 |
| CTCCC | 629755 | 1.0158957 | 5.744453 | 1 |
| CCGGG | 364960 | 0.9640211 | 5.0937095 | 1 |
| GTCCT | 875985 | 0.94009846 | 10.439948 | 1 |
| TCGTA | 1324215 | 0.93392694 | 7.7306905 | 7 |
| GTACA | 1239665 | 0.88232404 | 7.730677 | 6 |
| ACAAA | 1852225 | 0.8558033 | 5.1393175 | 8 |
| TACAA | 1746300 | 0.79952073 | 5.554891 | 7 |