![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-35-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 15554492 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
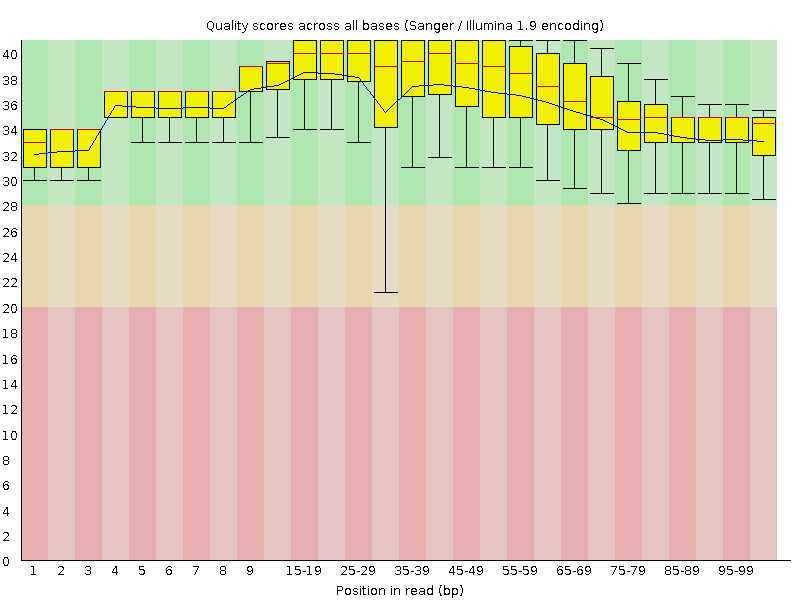
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
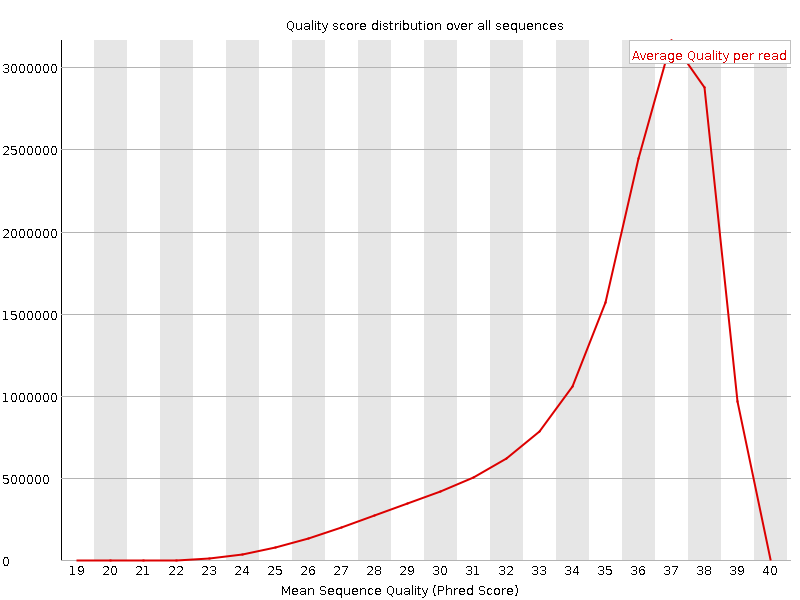
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
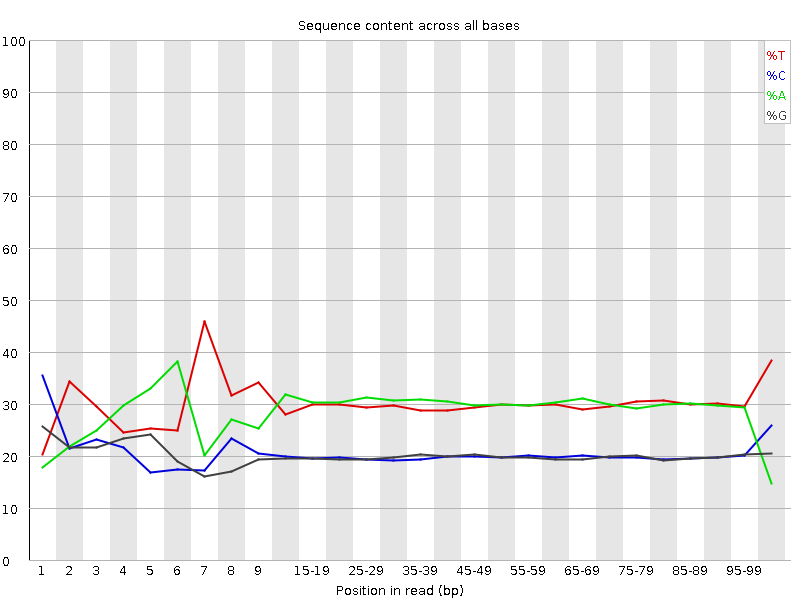
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
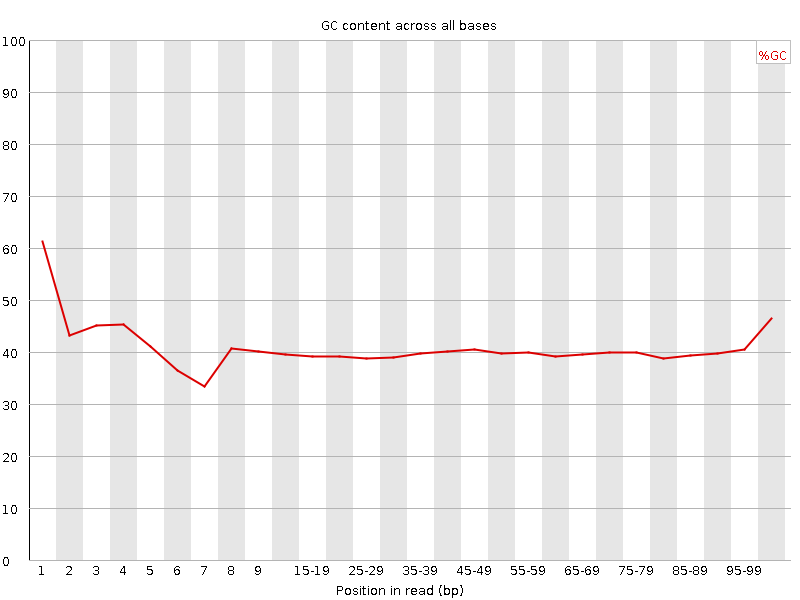
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
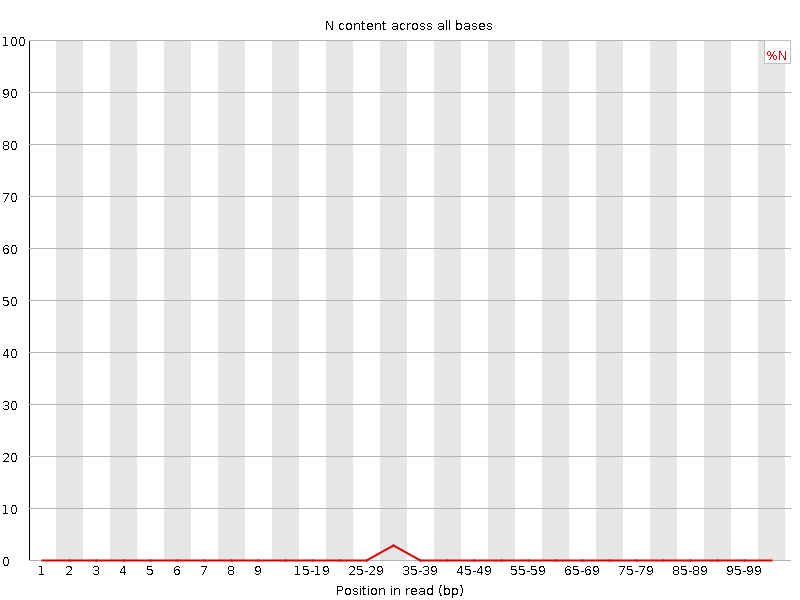
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
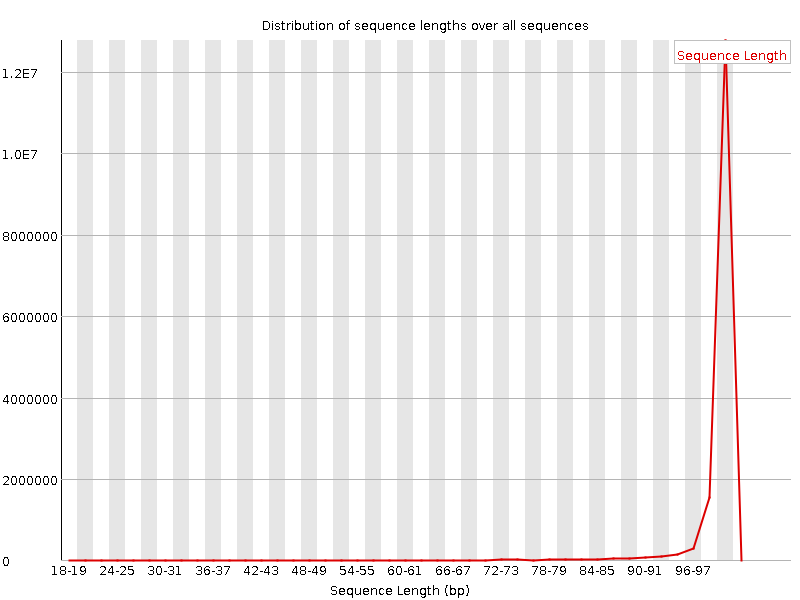
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 70246 | 0.45161230594994684 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 69003 | 0.44362104529032514 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 47739 | 0.3069145556151882 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 44867 | 0.28845043605409937 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 34086 | 0.21913926857913454 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 29552 | 0.18999013275393373 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 27868 | 0.1791636782480585 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 25414 | 0.16338688528047074 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 21201 | 0.136301461982815 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 20499 | 0.13178829626837057 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 18913 | 0.1215918848394406 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 16729 | 0.10755092483894683 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 16011 | 0.10293489494867464 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
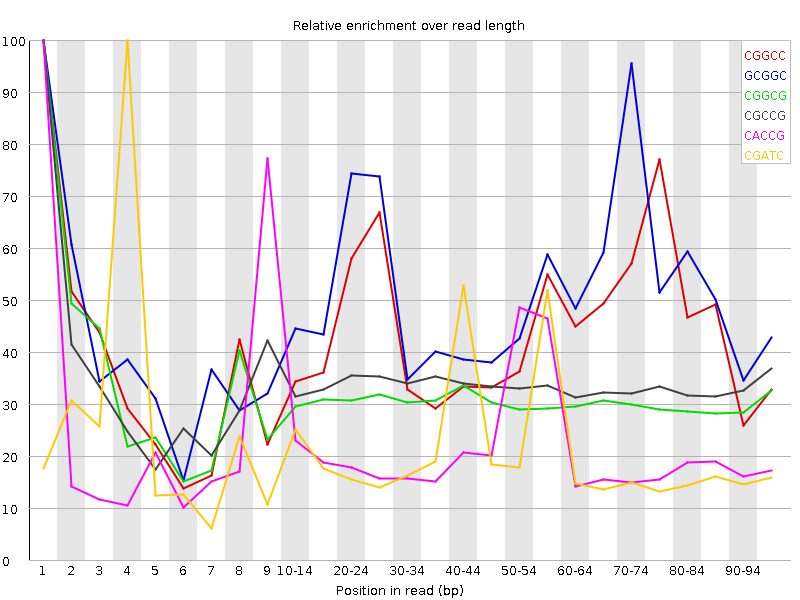
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1412460 | 2.9337564 | 6.6517096 | 1 |
| GCGGC | 1374255 | 2.876106 | 5.631816 | 1 |
| CGGCG | 1183190 | 2.476236 | 8.007785 | 1 |
| CGCCG | 1169275 | 2.428648 | 7.198587 | 1 |
| CACCG | 1639275 | 2.281651 | 10.368885 | 1 |
| CGATC | 2422540 | 2.2702863 | 10.655415 | 4 |
| GGTCG | 1564320 | 2.2043242 | 5.1271276 | 95-97 |
| CCTCG | 1581685 | 2.195283 | 5.76447 | 1 |
| CTGGC | 1475960 | 2.0641189 | 8.0050535 | 1 |
| GCCGG | 977805 | 2.0463967 | 5.221009 | 1 |
| CCGGC | 974135 | 2.0233316 | 6.844783 | 1 |
| CGACG | 1415300 | 1.9848864 | 5.352243 | 1 |
| CCGAT | 2022480 | 1.8953696 | 9.076769 | 3 |
| CGCGG | 879125 | 1.8398746 | 5.537015 | 1 |
| CCGCG | 882790 | 1.8336031 | 5.414866 | 1 |
| GGCTT | 1931500 | 1.8187257 | 5.4947896 | 3 |
| GCCGA | 1267635 | 1.7777938 | 5.44952 | 1 |
| GACCT | 1892040 | 1.7731277 | 6.9756727 | 95-97 |
| TGGCT | 1814940 | 1.7089711 | 5.262238 | 45-49 |
| ACCGA | 1805755 | 1.6970533 | 7.0691066 | 2 |
| CAGCG | 1172945 | 1.6449958 | 6.0980363 | 1 |
| GTCGC | 1098680 | 1.5364958 | 5.7885375 | 95-97 |
| GGGGC | 708970 | 1.495048 | 7.498348 | 95-97 |
| GATCT | 2362360 | 1.4906238 | 7.0126553 | 5 |
| CTTTC | 2323795 | 1.4511194 | 7.6309586 | 1 |
| TCCTT | 2320105 | 1.4488152 | 6.9324536 | 2 |
| CGAAC | 1500205 | 1.4098967 | 5.044592 | 90-94 |
| CTAAA | 3202295 | 1.357875 | 5.7428665 | 8 |
| CCTTT | 2146735 | 1.3405524 | 6.914217 | 3 |
| GCGTA | 1267370 | 1.1967483 | 5.001419 | 1 |
| AGACC | 1251715 | 1.176365 | 5.441806 | 95-97 |
| CAGAC | 1223190 | 1.1495571 | 5.159599 | 95-97 |
| TTTCG | 1781255 | 1.1207819 | 7.0506682 | 5 |
| TTCGT | 1769175 | 1.1131811 | 7.081198 | 6 |
| CGTAC | 1175655 | 1.1017666 | 9.312506 | 8 |
| GTCCT | 1021675 | 0.9547628 | 9.280695 | 1 |
| TCGTA | 1438570 | 0.90772223 | 6.986796 | 7 |
| GTACA | 1391665 | 0.88061 | 6.6986504 | 6 |
| TACAA | 1911010 | 0.810329 | 5.108077 | 7 |