![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-38-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 19235179 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
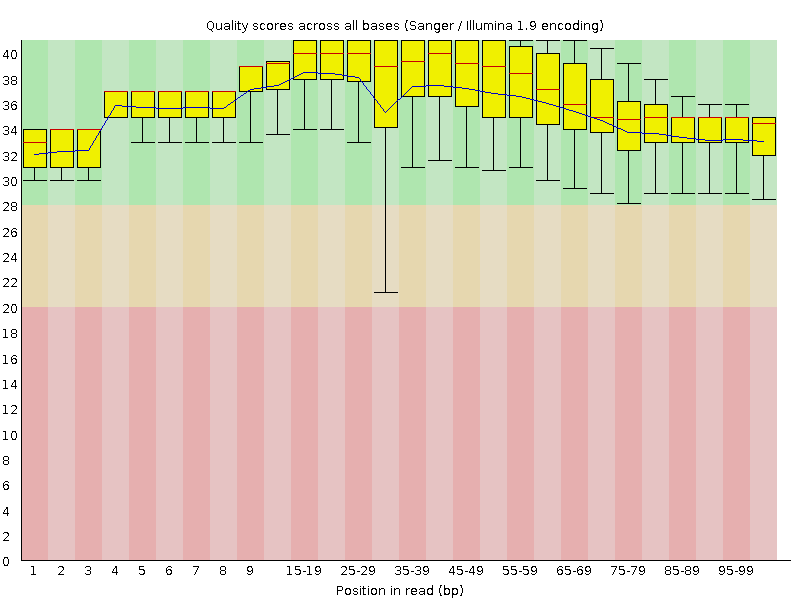
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

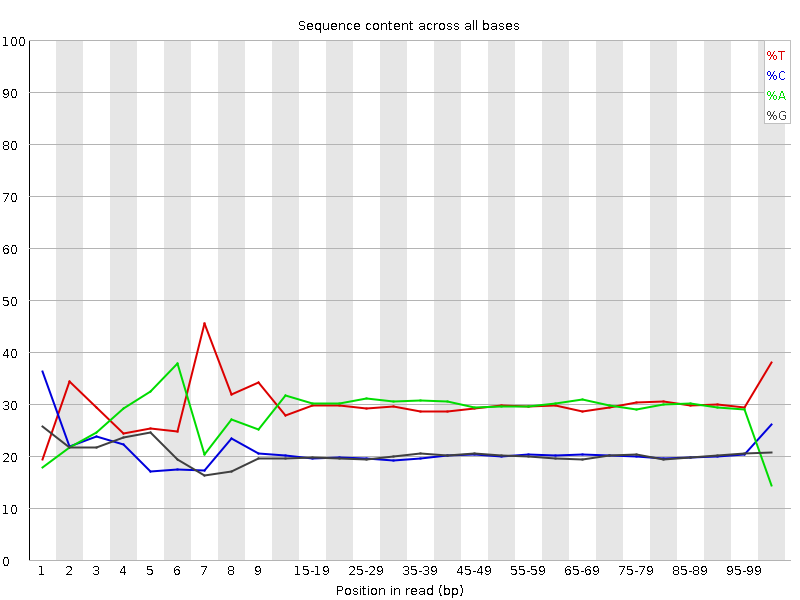
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
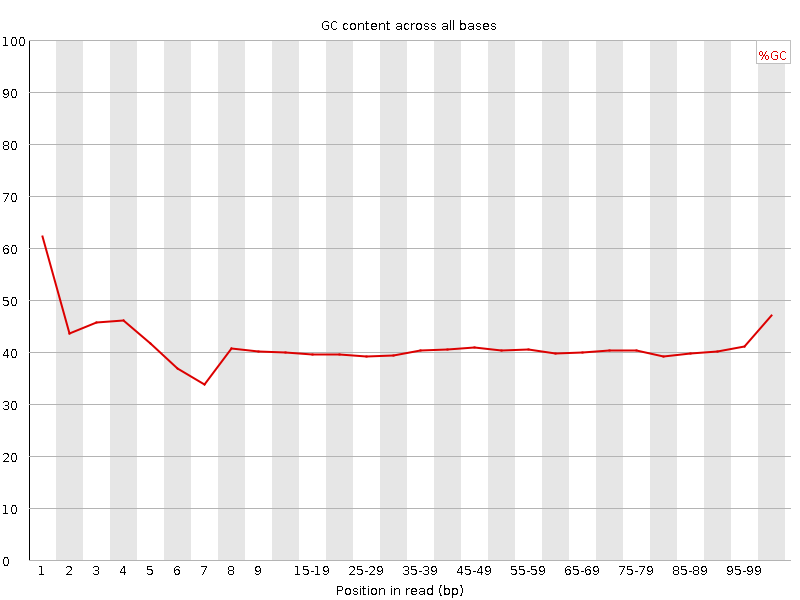
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
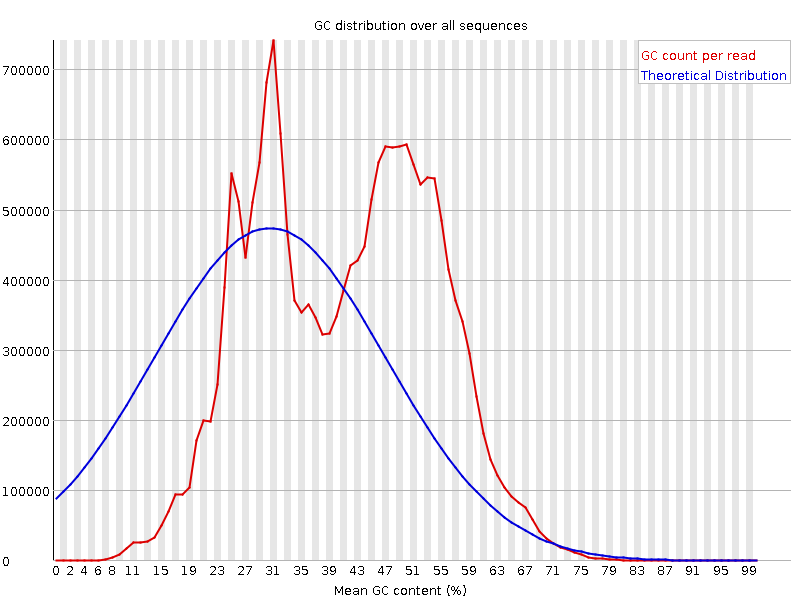
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
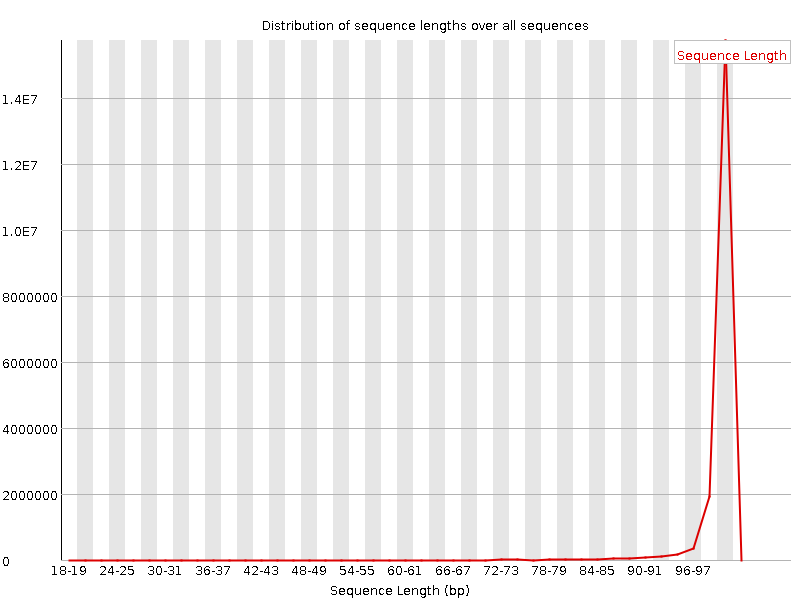
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels

![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 98632 | 0.5127688180078802 | No Hit |
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 95258 | 0.49522804024854666 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 67157 | 0.3491363402440913 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 54046 | 0.2809747702373864 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 45540 | 0.23675371048015723 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 40037 | 0.20814467076183696 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 38205 | 0.1986204547407643 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 33850 | 0.17597964645922973 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 29679 | 0.15429541882609982 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 28867 | 0.150073986834227 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 23324 | 0.12125699480103616 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 20147 | 0.10474038219244021 | No Hit |
| AATTAATTACCTTAGGGATAACAGCGTAATATTTTTAGAAAGATCATATT | 20087 | 0.10442845372013435 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 20006 | 0.10400735028252141 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
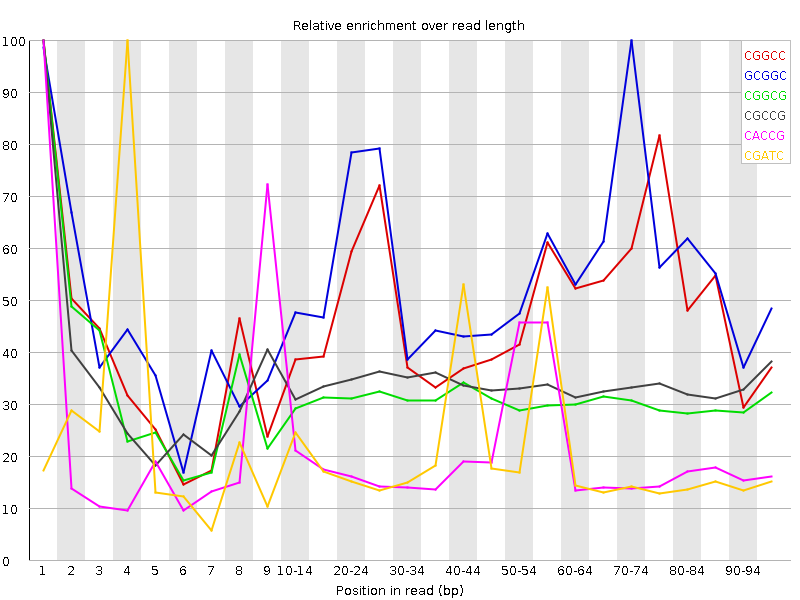
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1811940 | 2.895022 | 6.025029 | 1 |
| GCGGC | 1726860 | 2.781819 | 5.0586977 | 70-74 |
| CGGCG | 1506195 | 2.4263473 | 7.7807775 | 1 |
| CGCCG | 1485810 | 2.3739486 | 7.008984 | 1 |
| CACCG | 2147345 | 2.3352816 | 11.374256 | 1 |
| CGATC | 3122355 | 2.3281612 | 11.242862 | 4 |
| GGTCG | 2072760 | 2.2893603 | 5.5420556 | 95-97 |
| CCTCG | 2083500 | 2.2637637 | 6.211658 | 1 |
| CTGGC | 1961885 | 2.1491907 | 8.411237 | 1 |
| GCCGG | 1320545 | 2.1272814 | 5.0142446 | 1 |
| CCGGC | 1312400 | 2.0968828 | 6.6051545 | 1 |
| CCGAT | 2620655 | 1.9540721 | 9.789328 | 3 |
| CGACG | 1775320 | 1.9466048 | 5.018919 | 1 |
| GACCT | 2509520 | 1.8712051 | 7.344724 | 95-97 |
| GGCTT | 2490820 | 1.8708417 | 5.7431498 | 3 |
| CGCGG | 1139165 | 1.8350942 | 5.3029 | 1 |
| CCGCG | 1145285 | 1.8298755 | 5.2211385 | 1 |
| AGGTC | 2359535 | 1.7738667 | 5.3070273 | 95-97 |
| ACCGA | 2350355 | 1.7541391 | 7.801714 | 2 |
| TGGCT | 2327870 | 1.7484511 | 5.654004 | 45-49 |
| GCCGA | 1585395 | 1.7383556 | 5.1016455 | 1 |
| CAGCG | 1505710 | 1.6509825 | 6.825461 | 1 |
| GATCT | 3061025 | 1.5649163 | 7.5434113 | 5 |
| GTCGC | 1387135 | 1.519568 | 6.1905584 | 95-97 |
| TCCTT | 2904490 | 1.4713994 | 7.8593087 | 2 |
| CTTTC | 2862655 | 1.450206 | 8.0763855 | 1 |
| CGAAC | 1937075 | 1.445696 | 5.436587 | 90-94 |
| GGGGC | 877205 | 1.4247433 | 7.0936713 | 95-97 |
| CTAAA | 4009585 | 1.396538 | 6.0393667 | 8 |
| CCTTT | 2657675 | 1.3463641 | 7.7002425 | 3 |
| CAAAT | 3691640 | 1.2857978 | 5.310557 | 9 |
| GCGTA | 1663750 | 1.250785 | 5.2537184 | 1 |
| AGACC | 1613660 | 1.2043219 | 5.7144585 | 95-97 |
| CAGAC | 1565245 | 1.1681883 | 5.4575415 | 95-97 |
| CGTAC | 1534335 | 1.1440657 | 10.408159 | 8 |
| TTCGT | 2230420 | 1.139229 | 7.9228635 | 6 |
| TTTCG | 2225465 | 1.1366981 | 7.875496 | 5 |
| GTCCT | 1270440 | 0.9464228 | 10.399684 | 1 |
| TCGTA | 1838940 | 0.9401383 | 7.8565164 | 7 |
| GTACA | 1755605 | 0.8983608 | 7.2099853 | 9 |
| TACAA | 2313665 | 0.80584925 | 5.3986044 | 7 |