![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | scahan_VGN_20140730_2014_ddRad1_R1.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 152946005 |
| Filtered Sequences | 0 |
| Sequence length | 140 |
| %GC | 45 |
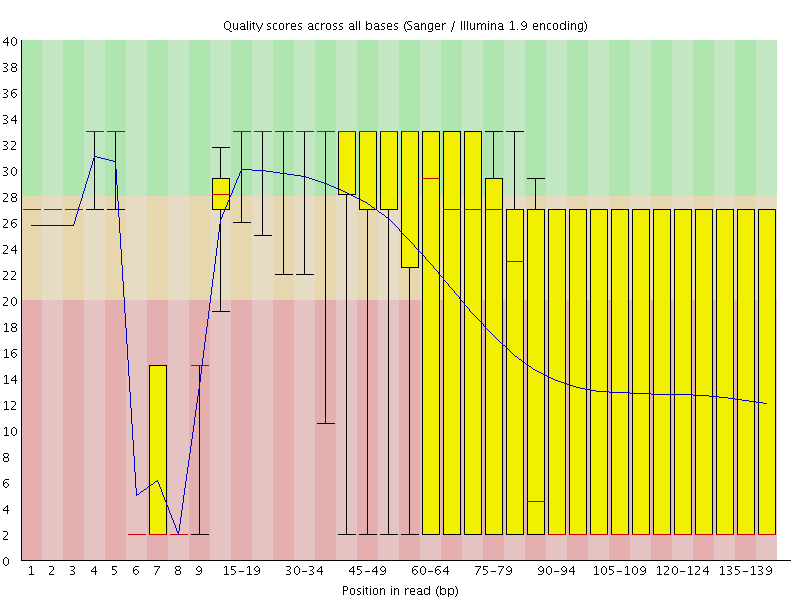
![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality
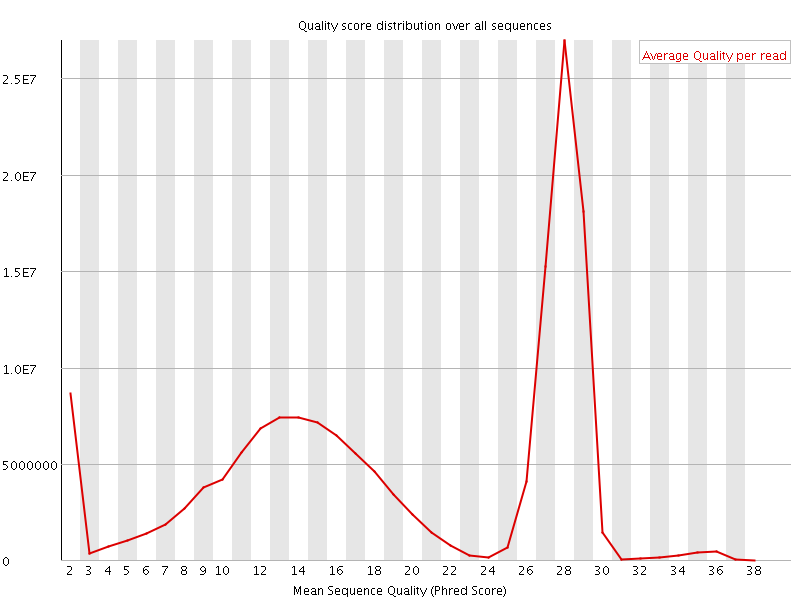
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
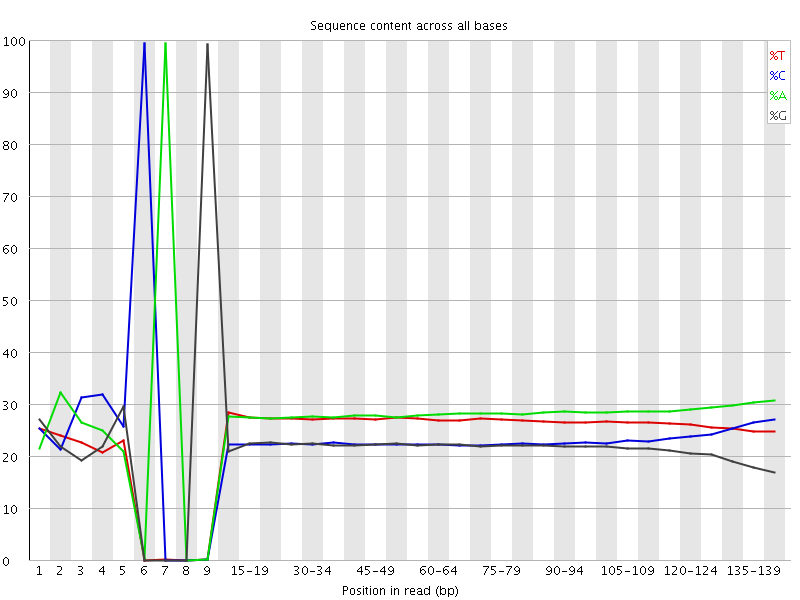
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
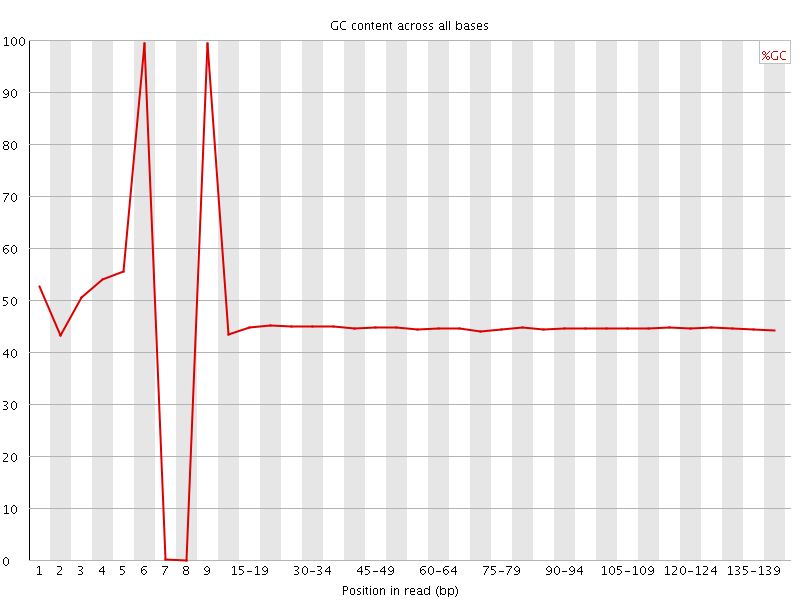
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
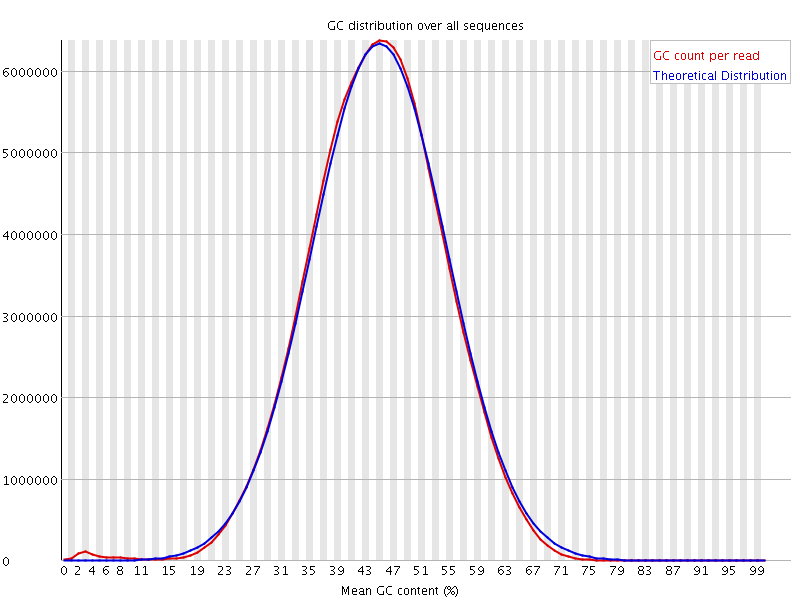
![[FAIL]](Icons/error.png) Per base N content
Per base N content
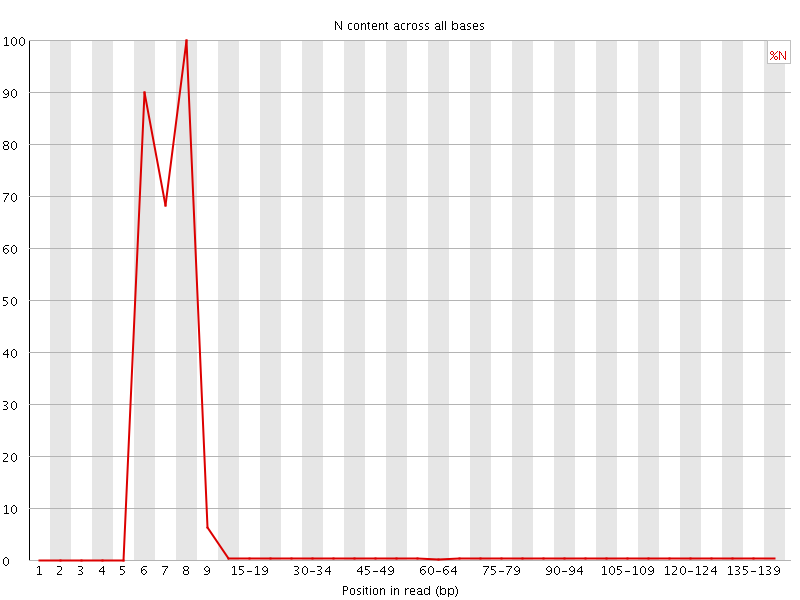
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
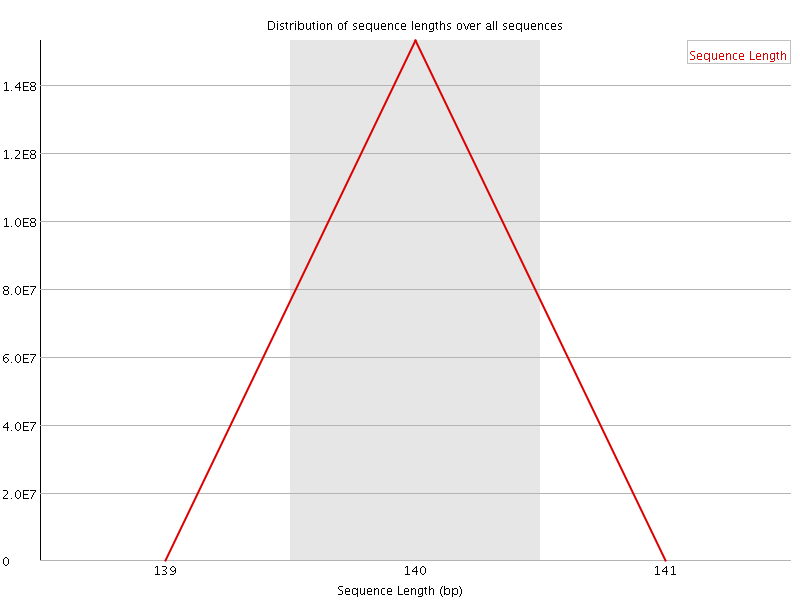
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
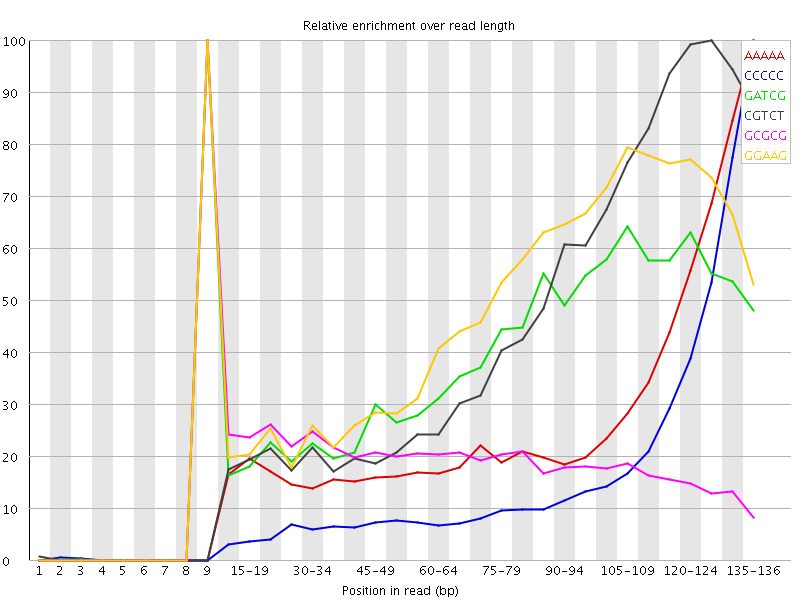
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 127309495 | 3.595933 | 13.304169 | 135-136 |
| CCCCC | 44054855 | 3.4812248 | 20.979788 | 135-136 |
| GATCG | 56667655 | 3.384793 | 8.526659 | 9 |
| CGTCT | 46818905 | 2.8864708 | 6.2691197 | 125-129 |
| GCGCG | 32522805 | 2.840213 | 14.331853 | 9 |
| GGAAG | 47593130 | 2.7541792 | 5.7055087 | 9 |
| CACAC | 50667725 | 2.6531065 | 18.871582 | 2 |
| GAAGA | 56298580 | 2.565151 | 5.1300697 | 9 |
| ACGTC | 43066320 | 2.488054 | 37.989292 | 2 |
| CGATC | 41722930 | 2.4104428 | 38.11918 | 1 |
| GCACA | 43611165 | 2.3609993 | 5.04747 | 9 |
| GAGCA | 41457290 | 2.3204591 | 33.680275 | 3 |
| AGATC | 49295105 | 2.318293 | 12.293492 | 2 |
| CGTCG | 29879145 | 2.1924143 | 32.869846 | 1 |
| TCACG | 35747265 | 2.065213 | 24.485823 | 1 |
| GAAAA | 56275885 | 2.0188568 | 7.146517 | 9 |
| GCCGT | 25355870 | 1.8605139 | 21.69288 | 1 |
| CTCCA | 31417115 | 1.7555511 | 21.471481 | 3 |
| GCGAG | 24635355 | 1.7513149 | 6.5356064 | 9 |
| CAGTC | 28771490 | 1.6622043 | 18.23334 | 2 |
| CCGTC | 23309390 | 1.6542854 | 21.491467 | 2 |
| GCGAT | 27630005 | 1.6503569 | 6.0173373 | 9 |
| GTTTT | 34391665 | 1.6000435 | 11.039002 | 9 |
| GCGTG | 21090750 | 1.6000049 | 8.501701 | 9 |
| GCCGC | 18520305 | 1.5643584 | 6.992747 | 9 |
| GCGGC | 17904305 | 1.5635811 | 6.0824194 | 9 |
| GTCGA | 25896695 | 1.5468253 | 37.47166 | 1 |
| TCGAT | 30720920 | 1.5417808 | 6.8806148 | 1 |
| GTGTG | 23359670 | 1.5394385 | 8.595813 | 9 |
| GTCGC | 20731740 | 1.5212135 | 32.79016 | 2 |
| TCTGC | 24612975 | 1.5174347 | 19.435463 | 1 |
| GTGCG | 19648310 | 1.4905772 | 9.5282755 | 9 |
| GCACG | 21632645 | 1.4874426 | 8.8926115 | 9 |
| CATCA | 32534790 | 1.479918 | 16.424852 | 3 |
| GCTCG | 20133945 | 1.4773498 | 6.600089 | 9 |
| GATTT | 33810740 | 1.4740404 | 10.261479 | 9 |
| GCGTT | 23087000 | 1.4715949 | 6.7947364 | 9 |
| GGAAA | 32280215 | 1.4707943 | 6.291256 | 9 |
| AATTA | 44763795 | 1.439877 | 9.197375 | 1 |
| GTTTC | 26779440 | 1.4342133 | 9.998888 | 9 |
| GCGAC | 20554270 | 1.4132944 | 7.253798 | 9 |
| CACGC | 21249735 | 1.4132179 | 28.423399 | 2 |
| GCGAA | 25177295 | 1.4092307 | 5.012575 | 9 |
| GTCCG | 18973115 | 1.3921726 | 43.66219 | 1 |
| GACGA | 24513535 | 1.3720784 | 5.16199 | 9 |
| AGTCA | 29097470 | 1.368421 | 27.256931 | 3 |
| CATAT | 34147845 | 1.3493336 | 36.823975 | 1 |
| GTATG | 25887810 | 1.3432554 | 5.373071 | 9 |
| TCCGC | 18831755 | 1.3365042 | 42.956356 | 2 |
| GCGTC | 18205165 | 1.3358233 | 5.146431 | 9 |
| CTGCG | 18096735 | 1.3278673 | 29.083755 | 1 |
| GGCGA | 18612225 | 1.3231337 | 6.282808 | 9 |
| TGCGC | 17941445 | 1.3164726 | 29.261667 | 2 |
| GCGCA | 19130220 | 1.315378 | 27.97045 | 3 |
| GAAAT | 34244595 | 1.3109891 | 6.589576 | 9 |
| TACGT | 26056330 | 1.3076804 | 32.683636 | 1 |
| ACACA | 30434795 | 1.2972873 | 15.4168 | 1 |
| GGCGG | 14306675 | 1.2917441 | 5.988195 | 9 |
| GTCGT | 20180595 | 1.286337 | 5.8656964 | 9 |
| ATATC | 32192600 | 1.2720731 | 36.934544 | 2 |
| GCCGA | 18379785 | 1.2637787 | 5.4547157 | 9 |
| GACGC | 18292080 | 1.257748 | 5.1986814 | 9 |
| GGCGC | 14391585 | 1.2568156 | 5.6905427 | 9 |
| GAGAT | 25799470 | 1.2544408 | 12.561747 | 1 |
| GATAT | 30587070 | 1.2495934 | 5.836399 | 9 |
| GACGT | 20795710 | 1.2421403 | 5.087642 | 9 |
| TCCCC | 18074265 | 1.2406958 | 5.1867723 | 135-136 |
| GTTGC | 19422075 | 1.2379879 | 23.48186 | 2 |
| GTTCG | 19421330 | 1.2379403 | 6.252055 | 9 |
| ATACG | 26300505 | 1.236883 | 25.755325 | 1 |
| CTTCC | 20724820 | 1.2358387 | 11.17977 | 2 |
| TACCG | 20845990 | 1.2043273 | 40.02076 | 1 |
| TCGAC | 20824400 | 1.20308 | 36.7662 | 2 |
| GCTGC | 16360285 | 1.2004533 | 5.475541 | 9 |
| TCGCA | 20710545 | 1.1965024 | 26.667393 | 3 |
| GCAAA | 26653810 | 1.174627 | 6.378541 | 9 |
| GCATT | 22976050 | 1.1530915 | 6.993327 | 9 |
| GCGCT | 15635535 | 1.147274 | 5.137965 | 9 |
| ACCGC | 17150860 | 1.1406214 | 46.576244 | 2 |
| GACAC | 21013625 | 1.1376249 | 32.260677 | 1 |
| GTATT | 26068435 | 1.1365008 | 6.590693 | 9 |
| CCGGC | 13438895 | 1.1351458 | 33.18079 | 2 |
| GCTTT | 20938575 | 1.1213969 | 6.0718937 | 9 |
| TTGCA | 22286080 | 1.1184641 | 18.999079 | 3 |
| ACCCC | 17328595 | 1.1146644 | 5.884208 | 135-136 |
| CGAAT | 23625705 | 1.1110902 | 15.287342 | 1 |
| GTTTG | 19864025 | 1.099903 | 6.9916 | 9 |
| GCATC | 18998230 | 1.0975775 | 20.516516 | 2 |
| CGACA | 20143785 | 1.0905341 | 35.556194 | 3 |
| GATGA | 22381955 | 1.0882719 | 5.2073956 | 9 |
| TCCGG | 14829390 | 1.0881222 | 28.487837 | 1 |
| ACGGT | 18192260 | 1.0866346 | 22.402601 | 1 |
| CGTAC | 18783525 | 1.0851734 | 14.458146 | 1 |
| GCGTA | 18062830 | 1.0789038 | 5.173581 | 9 |
| GCTTC | 17459230 | 1.0763934 | 5.892316 | 9 |
| GTTAT | 24688955 | 1.0763599 | 6.081671 | 9 |
| GTAAA | 27988235 | 1.0714763 | 5.925165 | 9 |
| GATTG | 20535885 | 1.065557 | 5.71338 | 9 |
| GTTTA | 24434880 | 1.065283 | 7.545501 | 9 |
| TGCAT | 21208490 | 1.0643836 | 17.73347 | 1 |
| ACGCA | 19627595 | 1.0625889 | 52.661648 | 3 |
| CGGCT | 14343595 | 1.0524765 | 18.544222 | 1 |
| TGGAA | 21520280 | 1.0463748 | 15.778442 | 1 |
| TATAC | 26399615 | 1.0431665 | 16.180195 | 1 |
| GTGCA | 17450175 | 1.0423095 | 7.178799 | 9 |
| CGGCA | 15084550 | 1.037201 | 27.076403 | 3 |
| GGGCG | 11426295 | 1.0316757 | 5.0782957 | 9 |
| GTACG | 17225125 | 1.0288671 | 5.576692 | 9 |
| ATACA | 27713155 | 1.0261672 | 7.5072455 | 3 |
| TACGC | 17753495 | 1.0256659 | 31.74366 | 2 |
| AAGGA | 22453920 | 1.0230755 | 7.3073363 | 1 |
| GTATC | 20321335 | 1.0198603 | 5.111257 | 9 |
| CAACC | 19435940 | 1.0177212 | 40.493843 | 1 |
| GTATA | 24425010 | 0.9978509 | 7.3725085 | 9 |
| GTGTA | 19145825 | 0.99343014 | 5.3166614 | 9 |
| CGTCA | 17115365 | 0.98879945 | 56.264442 | 3 |
| CTGTC | 15966225 | 0.9843468 | 8.092184 | 1 |
| GTGTT | 17744385 | 0.9825351 | 5.1152635 | 9 |
| CGGTC | 13195890 | 0.9682625 | 27.720083 | 2 |
| GATCC | 16738190 | 0.96700895 | 38.20638 | 2 |
| GCTGA | 16148800 | 0.9645776 | 17.860922 | 1 |
| GAATC | 20329395 | 0.95606846 | 15.346294 | 2 |
| GGCTC | 13011090 | 0.95470256 | 26.179497 | 1 |
| GTTCT | 17804170 | 0.9535292 | 5.22182 | 9 |
| AATCA | 25613000 | 0.9484023 | 12.264807 | 3 |
| TTCCA | 19455905 | 0.94441956 | 8.960221 | 3 |
| AACCA | 21888540 | 0.933002 | 31.790865 | 1 |
| ATGCA | 19796085 | 0.9309876 | 33.553097 | 3 |
| GTGAT | 17939395 | 0.93083155 | 6.6265135 | 9 |
| GGCGT | 12202805 | 0.9257398 | 5.271512 | 9 |
| TTACA | 23288410 | 0.9202289 | 11.343354 | 3 |
| ACACC | 17465950 | 0.91456693 | 31.60973 | 2 |
| TTACC | 18805000 | 0.9128236 | 25.928665 | 1 |
| CCGCA | 13708590 | 0.9116926 | 87.764755 | 3 |
| GCATA | 19323060 | 0.9087418 | 5.204838 | 9 |
| ACTTC | 18719490 | 0.90867287 | 9.106089 | 1 |
| GGTTG | 13782675 | 0.90829974 | 24.07205 | 1 |
| GATAC | 19134610 | 0.899879 | 9.334819 | 2 |
| GGAAC | 15882285 | 0.8889678 | 18.445732 | 2 |
| CTTGG | 13934635 | 0.8882114 | 20.492191 | 1 |
| GTACA | 18870590 | 0.8874626 | 18.584995 | 3 |
| ATTAC | 22427475 | 0.88620955 | 12.907913 | 1 |
| GGATA | 18220340 | 0.88592285 | 9.552854 | 1 |
| TTGGC | 13834715 | 0.8818424 | 20.634388 | 2 |
| TGTCC | 14290160 | 0.88101435 | 8.089634 | 2 |
| GCCAC | 13197990 | 0.8777351 | 21.237577 | 2 |
| CTGCC | 12327330 | 0.87488014 | 22.688797 | 2 |
| CTGAT | 17297360 | 0.868097 | 16.095695 | 1 |
| GAACA | 19696150 | 0.86800456 | 14.737261 | 3 |
| GGCAG | 12173225 | 0.8653884 | 5.103814 | 9 |
| GAGTC | 14361970 | 0.8578491 | 22.616661 | 1 |
| GGCCA | 12444455 | 0.85567033 | 21.71302 | 1 |
| GTAAC | 18120785 | 0.85220015 | 5.591344 | 9 |
| GCCTC | 12006335 | 0.8520989 | 6.621246 | 9 |
| TATCA | 21174230 | 0.8366882 | 37.00815 | 3 |
| GCTCC | 11749495 | 0.8338707 | 25.992111 | 2 |
| AACCC | 15786670 | 0.82663506 | 40.52792 | 2 |
| CTGGC | 11191355 | 0.8211776 | 17.54961 | 2 |
| ATGAG | 16598335 | 0.80705655 | 28.507372 | 1 |
| TGACA | 17045475 | 0.8016295 | 14.512876 | 3 |
| ATCCA | 17546355 | 0.79813534 | 30.58466 | 3 |
| TGATC | 15304745 | 0.7680943 | 16.09289 | 2 |
| ACCAC | 14647885 | 0.767005 | 39.15904 | 2 |
| TGGCA | 12829535 | 0.7663159 | 33.525524 | 3 |
| CACCA | 14593075 | 0.764135 | 31.49576 | 3 |
| TGCCA | 13168715 | 0.7607912 | 18.5851 | 3 |
| TCAGT | 15118450 | 0.75874466 | 15.729582 | 1 |
| GGACA | 13479350 | 0.75447 | 9.137747 | 3 |
| GATCA | 15586265 | 0.73300433 | 34.021904 | 3 |
| GACTT | 14402010 | 0.72278893 | 5.5220957 | 9 |
| GTCCA | 12480975 | 0.7210586 | 30.415018 | 3 |
| ATACC | 15612715 | 0.7101794 | 18.701698 | 2 |
| GTACC | 12128355 | 0.7006868 | 14.501401 | 2 |
| GCTAC | 11949405 | 0.6903484 | 19.51723 | 2 |
| CTGAC | 11931540 | 0.6893163 | 17.653685 | 2 |
| GTAGT | 13029940 | 0.67609185 | 13.078099 | 1 |
| TGAGC | 11229050 | 0.670718 | 35.245613 | 2 |
| CCACA | 12806400 | 0.6705796 | 55.136494 | 3 |
| ACTGG | 11203360 | 0.66918343 | 14.103056 | 1 |
| AGGAC | 11653275 | 0.65226036 | 9.122763 | 2 |
| TAGTA | 15253130 | 0.62314606 | 14.185398 | 1 |
| TACCC | 10871030 | 0.6074602 | 30.095179 | 2 |
| AGCTA | 12882425 | 0.60584587 | 15.780735 | 1 |
| GGTCA | 10132170 | 0.6052007 | 22.940031 | 3 |
| GCTCA | 10466785 | 0.6046936 | 14.587078 | 3 |
| TACCA | 13145690 | 0.5979612 | 44.83647 | 3 |
| AGTAC | 12433895 | 0.58475196 | 16.45458 | 2 |
| CTACA | 12454385 | 0.5665157 | 15.189374 | 3 |
| ACCCA | 10615910 | 0.5558793 | 68.81912 | 3 |
| AGTCC | 8555820 | 0.49429208 | 22.157745 | 2 |
| TAGTC | 9375655 | 0.47053295 | 12.777428 | 2 |
| GCATG | 7104715 | 0.42436892 | 42.057972 | 1 |
| CATGC | 2075565 | 0.11991082 | 40.98566 | 2 |